An analogue of the Prolactin Releasing Peptide reduces obesity and promotes adult neurogenesis
- PMID: 38177913
- PMCID: PMC10897398
- DOI: 10.1038/s44319-023-00016-2
An analogue of the Prolactin Releasing Peptide reduces obesity and promotes adult neurogenesis
Abstract
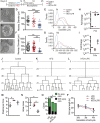
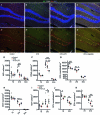
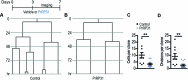
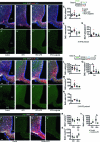
Hypothalamic Adult Neurogenesis (hAN) has been implicated in regulating energy homeostasis. Adult-generated neurons and adult Neural Stem Cells (aNSCs) in the hypothalamus control food intake and body weight. Conversely, diet-induced obesity (DIO) by high fat diets (HFD) exerts adverse influence on hAN. However, the effects of anti-obesity compounds on hAN are not known. To address this, we administered a lipidized analogue of an anti-obesity neuropeptide, Prolactin Releasing Peptide (PrRP), so-called LiPR, to mice. In the HFD context, LiPR rescued the survival of adult-born hypothalamic neurons and increased the number of aNSCs by reducing their activation. LiPR also rescued the reduction of immature hippocampal neurons and modulated calcium dynamics in iPSC-derived human neurons. In addition, some of these neurogenic effects were exerted by another anti-obesity compound, Liraglutide. These results show for the first time that anti-obesity neuropeptides influence adult neurogenesis and suggest that the neurogenic process can serve as a target of anti-obesity pharmacotherapy.
Keywords: Adult neurogenesis; Anti-obesity peptides; Hypothalamus; Neural stem cells; Prolactin Releasing Peptide.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Repeated peripheral administration of lipidized prolactin-releasing peptide analog induces c-fos and FosB expression in neurons of dorsomedial hypothalamic nucleus in male C57 mice.Neurochem Int. 2018 Jun;116:77-84. doi: 10.1016/j.neuint.2018.03.013. Epub 2018 Mar 27. Neurochem Int. 2018. PMID: 29601847
-
Prolactin-releasing peptide: a new tool for obesity treatment.J Endocrinol. 2016 Aug;230(2):R51-8. doi: 10.1530/JOE-16-0046. J Endocrinol. 2016. PMID: 27418033 Review.
-
Palmitoylated PrRP analog decreases body weight in DIO rats but not in ZDF rats.J Endocrinol. 2016 May;229(2):85-96. doi: 10.1530/JOE-15-0519. Epub 2016 Feb 23. J Endocrinol. 2016. PMID: 26906745
-
Novel lipidized analogs of prolactin-releasing peptide have prolonged half-lives and exert anti-obesity effects after peripheral administration.Int J Obes (Lond). 2015 Jun;39(6):986-93. doi: 10.1038/ijo.2015.28. Epub 2015 Mar 16. Int J Obes (Lond). 2015. PMID: 25771926
-
Physiological roles of prolactin-releasing peptide.Regul Pept. 2005 Mar 15;126(1-2):27-33. doi: 10.1016/j.regpep.2004.08.008. Regul Pept. 2005. PMID: 15620410 Review.
Cited by
-
Anorexigenic neuropeptides as anti-obesity and neuroprotective agents: exploring the neuroprotective effects of anorexigenic neuropeptides.Biosci Rep. 2024 Apr 24;44(4):BSR20231385. doi: 10.1042/BSR20231385. Biosci Rep. 2024. PMID: 38577975 Free PMC article. Review.
References
-
- Andrews S (2010). FastQC: a quality control tool for high throughput sequence data., Babraham Bioinformatics Institute
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

