Colorectal cancer with low SLC35A3 is associated with immune infiltrates and poor prognosis
- PMID: 38172565
- PMCID: PMC10764849
- DOI: 10.1038/s41598-023-51028-w
Colorectal cancer with low SLC35A3 is associated with immune infiltrates and poor prognosis
Abstract
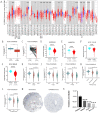
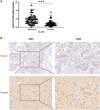
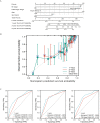
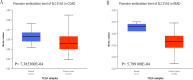
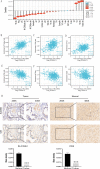
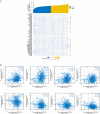
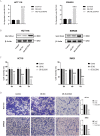
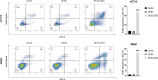
The expression level of SLC35A3 is associated with the prognosis of many cancers, but its role in colorectal cancer (CRC) is unclear. The purpose of our study was to elucidate the role of SLC35A3 in CRC. The expression levels of SLC35A3 in CRC were evaluated through tumor immune resource assessment (TIMER), The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), International Cancer Genome Consortium (ICGC), Human Protein Atlas (HPA), qRT-PCR, and immunohistochemical evaluation. TCGA, GEO, and ICGC databases were used to analyze the diagnostic and prognostic value of SLC35A3 in CRC. A overall survival (OS) model was constructed and validated based on the expression level of SLC35A3 and multivariable analysis results. The cBioPortal tool was used to analyze SLC35A3 mutation in CRC. The UALCAN tool was used to analyze the promoter methylation level of SLC35A3 in colorectal cancer. In addition, the role of SLC35A3 in CRC was determined through GO analysis, KEGG analysis, gene set enrichment analysis (GSEA), immune infiltration analysis, and immune checkpoint correlation analysis. In vitro experiments validated the function of SLC35A3 in colorectal cancer cells. Compared with adjacent normal tissues and colonic epithelial cells, the expression of SLC35A3 was decreased in CRC tissues and CRC cell lines. Low expression of SLC35A3 was associated with N stage, pathological stage, and lymphatic infiltration, and it was unfavorable for OS, disease-specific survival (DSS), recurrence-free survival (RFS), and post-progression survival (PPS). According to the Receiver Operating Characteristic (ROC) analysis, SLC35A3 is a potential important diagnostic biomarker for CRC patients. The nomograph based on the expression level of SLC35A3 showed a better predictive model for OS than single prognostic factors and TNM staging. SLC35A3 has multiple types of mutations in CRC, and its promoter methylation level is significantly decreased. GO and KEGG analysis indicated that SLC35A3 may be involved in transmembrane transport protein activity, cell communication, and interaction with neurotransmitter receptors. GSEA revealed that SLC35A3 may be involved in energy metabolism, DNA repair, and cancer pathways. In addition, SLC35A3 was closely related to immune cell infiltration and immune checkpoint expression. Immunohistochemistry confirmed the positive correlation between SLC35A3 and helper T cell infiltration. In vitro experiments showed that overexpression of SLC35A3 inhibited the proliferation and invasion capability of colorectal cancer cells and promoted apoptosis. The results of this study indicate that decreased expression of SLC35A3 is closely associated with poor prognosis and immune cell infiltration in colorectal cancer, and it can serve as a promising independent prognostic biomarker and potential therapeutic target.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
UBR1 is a prognostic biomarker and therapeutic target associated with immune cell infiltration in gastric cancer.Aging (Albany NY). 2024 Aug 23;16(16):12029-12049. doi: 10.18632/aging.206079. Epub 2024 Aug 23. Aging (Albany NY). 2024. PMID: 39181686 Free PMC article.
-
Decreased E2F2 Expression Correlates with Poor Prognosis and Immune Infiltrates in Patients with Colorectal Cancer.J Cancer. 2022 Jan 1;13(2):653-668. doi: 10.7150/jca.61415. eCollection 2022. J Cancer. 2022. PMID: 35069909 Free PMC article.
-
Tumor microenvironment-related gene selenium-binding protein 1 (SELENBP1) is associated with immunotherapy efficacy and survival in colorectal cancer.BMC Gastroenterol. 2022 Oct 17;22(1):437. doi: 10.1186/s12876-022-02532-2. BMC Gastroenterol. 2022. PMID: 36253721 Free PMC article.
-
Novel biomarkers for the diagnosis and prognosis of colorectal cancer.Intest Res. 2020 Apr;18(2):168-183. doi: 10.5217/ir.2019.00080. Epub 2019 Nov 30. Intest Res. 2020. PMID: 31766836 Free PMC article. Review.
-
Systems biology and OMIC data integration to understand gastrointestinal cancers.World J Clin Oncol. 2022 Oct 24;13(10):762-778. doi: 10.5306/wjco.v13.i10.762. World J Clin Oncol. 2022. PMID: 36337313 Free PMC article. Review.
Cited by
-
TOX: a potential new immune checkpoint in cancers by pancancer analysis.Discov Oncol. 2024 Aug 16;15(1):354. doi: 10.1007/s12672-024-01236-1. Discov Oncol. 2024. PMID: 39152366 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

