Blocking methionine catabolism induces senescence and confers vulnerability to GSK3 inhibition in liver cancer
- PMID: 38168934
- PMCID: PMC11277537
- DOI: 10.1038/s43018-023-00671-3
Blocking methionine catabolism induces senescence and confers vulnerability to GSK3 inhibition in liver cancer
Abstract
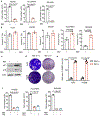
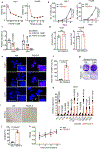
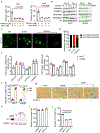
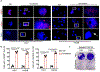
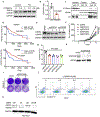
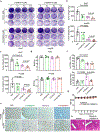
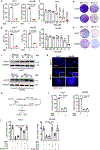
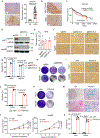
Availability of the essential amino acid methionine affects cellular metabolism and growth, and dietary methionine restriction has been implicated as a cancer therapeutic strategy. Nevertheless, how liver cancer cells respond to methionine deprivation and underlying mechanisms remain unclear. Here we find that human liver cancer cells undergo irreversible cell cycle arrest upon methionine deprivation in vitro. Blocking methionine adenosyl transferase 2A (MAT2A)-dependent methionine catabolism induces cell cycle arrest and DNA damage in liver cancer cells, resulting in cellular senescence. A pharmacological screen further identified GSK3 inhibitors as senolytics that selectively kill MAT2A-inhibited senescent liver cancer cells. Importantly, combined treatment with MAT2A and GSK3 inhibitors therapeutically blunts liver tumor growth in vitro and in vivo across multiple models. Together, methionine catabolism is essential for liver tumor growth, and its inhibition can be exploited as an improved pro-senescence strategy for combination with senolytic agents to treat liver cancer.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no potential conflicts of interest.
Figures





Similar articles
-
Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis.J Hepatol. 2013 Oct;59(4):830-41. doi: 10.1016/j.jhep.2013.04.031. Epub 2013 May 7. J Hepatol. 2013. PMID: 23665184 Review.
-
Role of promoter methylation in increased methionine adenosyltransferase 2A expression in human liver cancer.Am J Physiol Gastrointest Liver Physiol. 2001 Feb;280(2):G184-90. doi: 10.1152/ajpgi.2001.280.2.G184. Am J Physiol Gastrointest Liver Physiol. 2001. PMID: 11208539
-
S-adenosylmethionine biosynthesis is a targetable metabolic vulnerability of cancer stem cells.Breast Cancer Res Treat. 2019 May;175(1):39-50. doi: 10.1007/s10549-019-05146-7. Epub 2019 Feb 2. Breast Cancer Res Treat. 2019. PMID: 30712196 Free PMC article.
-
Overview of Methionine Adenosyltransferase 2A (MAT2A) as an Anticancer Target: Structure, Function, and Inhibitors.J Med Chem. 2022 Jul 28;65(14):9531-9547. doi: 10.1021/acs.jmedchem.2c00395. Epub 2022 Jul 7. J Med Chem. 2022. PMID: 35796517 Review.
-
Role of methionine adenosyltransferase 2A and S-adenosylmethionine in mitogen-induced growth of human colon cancer cells.Gastroenterology. 2007 Jul;133(1):207-18. doi: 10.1053/j.gastro.2007.03.114. Epub 2007 Apr 11. Gastroenterology. 2007. PMID: 17631143
Cited by
-
Harnessing CD8+ T cell dynamics in hepatitis B virus-associated liver diseases: Insights, therapies and future directions.Clin Transl Med. 2024 Jul;14(7):e1731. doi: 10.1002/ctm2.1731. Clin Transl Med. 2024. PMID: 38935536 Free PMC article. Review.
-
Role of Folate in Liver Diseases.Nutrients. 2024 Jun 14;16(12):1872. doi: 10.3390/nu16121872. Nutrients. 2024. PMID: 38931227 Free PMC article. Review.
-
Rescue of Methionine Dependence by Cobalamin in a Human Colorectal Cancer Cell Line.Nutrients. 2024 Mar 28;16(7):997. doi: 10.3390/nu16070997. Nutrients. 2024. PMID: 38613029 Free PMC article.
-
Building a translational cancer dependency map for The Cancer Genome Atlas.Nat Cancer. 2024 Aug;5(8):1176-1194. doi: 10.1038/s43018-024-00789-y. Epub 2024 Jul 15. Nat Cancer. 2024. PMID: 39009815 Free PMC article.
-
Roles of chromatin and genome instability in cellular senescence and their relevance to ageing and related diseases.Nat Rev Mol Cell Biol. 2024 Dec;25(12):979-1000. doi: 10.1038/s41580-024-00775-3. Epub 2024 Oct 3. Nat Rev Mol Cell Biol. 2024. PMID: 39363000 Review.
References
-
- Siegel RL, Miller KD, Fuchs HE & Jemal A. Cancer Statistics, 2021. CA Cancer J Clin 71, 7–33 (2021). - PubMed
-
- Finn RS et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med 382, 1894–1905 (2020). - PubMed
-
- Hanahan D. & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). - PubMed
-
- DeBerardinis RJ & Chandel NS We need to talk about the Warburg effect. Nat Metab 2, 127–129 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

