Higher-order protein assembly controls kinetochore formation
- PMID: 38168769
- PMCID: PMC10842828
- DOI: 10.1038/s41556-023-01313-7
Higher-order protein assembly controls kinetochore formation
Abstract
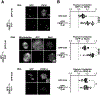
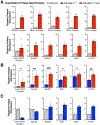
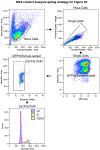
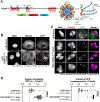
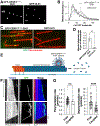
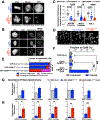
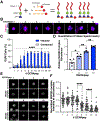
To faithfully segregate chromosomes during vertebrate mitosis, kinetochore-microtubule interactions must be restricted to a single site on each chromosome. Prior work on pair-wise kinetochore protein interactions has been unable to identify the mechanisms that prevent outer kinetochore formation in regions with a low density of CENP-A nucleosomes. To investigate the impact of higher-order assembly on kinetochore formation, we generated oligomers of the inner kinetochore protein CENP-T using two distinct, genetically engineered systems in human cells. Although individual CENP-T molecules interact poorly with outer kinetochore proteins, oligomers that mimic centromeric CENP-T density trigger the robust formation of functional, cytoplasmic kinetochore-like particles. Both in cells and in vitro, each molecule of oligomerized CENP-T recruits substantially higher levels of outer kinetochore components than monomeric CENP-T molecules. Our work suggests that the density dependence of CENP-T restricts outer kinetochore recruitment to centromeres, where densely packed CENP-A recruits a high local concentration of inner kinetochore proteins.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests Statement
The authors have no competing interests.
Figures








Similar articles
-
A super-resolution map of the vertebrate kinetochore.Proc Natl Acad Sci U S A. 2010 Jun 8;107(23):10484-9. doi: 10.1073/pnas.1002325107. Epub 2010 May 18. Proc Natl Acad Sci U S A. 2010. PMID: 20483991 Free PMC article.
-
An evolutionary perspective on the relationship between kinetochore size and CENP-E dependence for chromosome alignment.J Cell Sci. 2024 Dec 15;137(24):jcs263466. doi: 10.1242/jcs.263466. Epub 2024 Dec 19. J Cell Sci. 2024. PMID: 39698944
-
Sgo1 interacts with CENP-A to guide accurate chromosome segregation in mitosis.J Mol Cell Biol. 2024 Apr 4;15(10):mjad061. doi: 10.1093/jmcb/mjad061. J Mol Cell Biol. 2024. PMID: 37777834 Free PMC article.
-
Posttranslational modifications of CENP-A: marks of distinction.Chromosoma. 2018 Sep;127(3):279-290. doi: 10.1007/s00412-018-0665-x. Epub 2018 Mar 22. Chromosoma. 2018. PMID: 29569072 Free PMC article. Review.
-
Conserved and divergent mechanisms of inner kinetochore assembly onto centromeric chromatin.Curr Opin Struct Biol. 2023 Aug;81:102638. doi: 10.1016/j.sbi.2023.102638. Epub 2023 Jun 20. Curr Opin Struct Biol. 2023. PMID: 37343495 Review.
Cited by
-
Bridging the gap: unravelling plant centromeres in the telomere-to-telomere era.New Phytol. 2024 Dec;244(6):2143-2149. doi: 10.1111/nph.20149. Epub 2024 Sep 27. New Phytol. 2024. PMID: 39329317 Free PMC article. Review.
-
The structure, function, and evolution of plant centromeres.Genome Res. 2024 Mar 20;34(2):161-178. doi: 10.1101/gr.278409.123. Genome Res. 2024. PMID: 38485193 Free PMC article. Review.
-
A noncanonical GTPase signaling mechanism controls exit from mitosis in budding yeast.Proc Natl Acad Sci U S A. 2024 Nov 5;121(45):e2413873121. doi: 10.1073/pnas.2413873121. Epub 2024 Oct 30. Proc Natl Acad Sci U S A. 2024. PMID: 39475649 Free PMC article.
-
Regulation of outer kinetochore assembly during meiosis I and II by CENP-A and KNL-2/M18BP1 in C. elegans oocytes.Curr Biol. 2024 Nov 4;34(21):4853-4868.e6. doi: 10.1016/j.cub.2024.09.004. Epub 2024 Sep 30. Curr Biol. 2024. PMID: 39353426
-
A noncanonical GTPase signaling mechanism controls exit from mitosis in budding yeast.bioRxiv [Preprint]. 2024 Jul 4:2024.05.16.594582. doi: 10.1101/2024.05.16.594582. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Nov 5;121(45):e2413873121. doi: 10.1073/pnas.2413873121 PMID: 38798491 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

