Trichinella spiralis -induced immunomodulation signatures on gut microbiota and metabolic pathways in mice
- PMID: 38166140
- PMCID: PMC10786400
- DOI: 10.1371/journal.ppat.1011893
Trichinella spiralis -induced immunomodulation signatures on gut microbiota and metabolic pathways in mice
Abstract
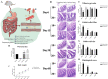
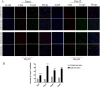
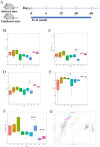
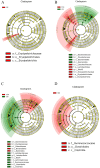
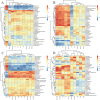
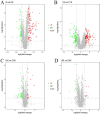
The hygiene hypothesis proposes that decreased exposure to infectious agents in developed countries may contribute to the development of allergic and autoimmune diseases. Trichinella spiralis, a parasitic roundworm, causes trichinellosis, also known as trichinosis, in humans. T. spiralis had many hosts, and almost any mammal could become infected. Adult worms lived in the small intestine, while the larvae lived in muscle cells of the same mammal. T. spiralis was a significant public health threat because it could cause severe illness and even death in humans who eat undercooked or raw meat containing the parasite. The complex interactions between gastrointestinal helminths, gut microbiota, and the host immune system present a challenge for researchers. Two groups of mice were infected with T. spiralis vs uninfected control, and the experiment was conducted over 60 days. The 16S rRNA gene sequences and untargeted LC/MS-based metabolomics of fecal and serum samples, respectively, from different stages of development of the Trichinella spiralis-mouse model, were examined in this study. Gut microbiota alterations and metabolic activity accompanied by parasite-induced immunomodulation were detected. The inflammation parameters of the duodenum (villus/crypt ratio, goblet cell number and size, and histological score) were involved in active inflammation and oxidative metabolite profiles. These profiles included increased biosynthesis of phenylalanine, tyrosine, and tryptophan while decreasing cholesterol metabolism and primary and secondary bile acid biosynthesis. These disrupted metabolisms adapted to infection stress during the enteral and parenteral phases and then return to homeostasis during the encapsulated phase. There was a shift from an abundance of Bacteroides in the parenteral phase to an abundance of probiotic Lactobacillus and Treg-associated-Clostridia in the encapsulated phase. Th2 immune response (IL-4/IL-5/IL-13), lamina propria Treg, and immune hyporesponsiveness metabolic pathways (decreased tropane, piperidine and pyridine alkaloid biosynthesis and biosynthesis of alkaloids derived from ornithine, lysine, and nicotinic acid) were all altered. These findings enhanced our understanding of gut microbiota and metabolic profiles of Trichinella -infected mice, which could be a driving force in parasite-shaping immune system maintenance.
Copyright: © 2024 Sun et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
The impact of MIF deficiency on alterations of fecal microbiota in C57BL/6 mice induced by Trichinella spiralis infection.FASEB J. 2023 Oct;37(10):e23202. doi: 10.1096/fj.202300179RR. FASEB J. 2023. PMID: 37732633
-
The gut microbiota contributes to changes in the host immune response induced by Trichinella spiralis.PLoS Negl Trop Dis. 2023 Aug 16;17(8):e0011479. doi: 10.1371/journal.pntd.0011479. eCollection 2023 Aug. PLoS Negl Trop Dis. 2023. PMID: 37585413 Free PMC article.
-
Higher mucosal type II immunity is associated with increased gut microbiota diversity in BALB/c mice after Trichinella spiralis infection.Mol Immunol. 2021 Oct;138:87-98. doi: 10.1016/j.molimm.2021.07.014. Epub 2021 Aug 4. Mol Immunol. 2021. PMID: 34364076
-
Trichinella spiralis immunomodulation: an interactive multifactorial process.Expert Rev Clin Immunol. 2013 Jul;9(7):669-75. doi: 10.1586/1744666X.2013.811187. Expert Rev Clin Immunol. 2013. PMID: 23899237 Review.
-
Trichinella spiralis: inflammation modulator.J Helminthol. 2020 Sep 21;94:e193. doi: 10.1017/S0022149X20000802. J Helminthol. 2020. PMID: 32951619 Review.
Cited by
-
The gut microbiota is essential for Trichinella spiralis-evoked suppression of colitis.PLoS Negl Trop Dis. 2024 Nov 4;18(11):e0012645. doi: 10.1371/journal.pntd.0012645. eCollection 2024 Nov. PLoS Negl Trop Dis. 2024. PMID: 39495798 Free PMC article.
-
Biological characteristics of a new long-chain fatty acid transport protein 1 from Trichinella spiralis and its participation in lipid metabolism, larval moulting, and development.Vet Res. 2024 Sep 30;55(1):126. doi: 10.1186/s13567-024-01380-0. Vet Res. 2024. PMID: 39350238 Free PMC article.
-
Modulation of the rat intestinal microbiota in the course of Anisakis pegreffii infection.Front Vet Sci. 2024 May 9;11:1403920. doi: 10.3389/fvets.2024.1403920. eCollection 2024. Front Vet Sci. 2024. PMID: 38784661 Free PMC article.
-
ACV with/without IVM: a new talk on intestinal CDX2 and muscular CD34 and Cyclin D1 during Trichinella spiralis infection.Helminthologia. 2024 Jul 16;61(2):124-141. doi: 10.2478/helm-2024-0013. eCollection 2024 Jun. Helminthologia. 2024. PMID: 39040803 Free PMC article.
References
-
- Jenkins SJ, Perona-Wright G, Worsley AG, Ishii N, MacDonald AS. Dendritic cell expression of OX40 ligand acts as a costimulatory, not polarizing, signal for optimal Th2 priming and memory induction in vivo. J Immunol. 2007;179(6):3515–23. Epub 2007/09/06. doi: 10.4049/jimmunol.179.6.3515 . - DOI - PubMed
-
- Ekkens MJ, Liu Z, Liu Q, Whitmire J, Xiao S, Foster A, et al.. The role of OX40 ligand interactions in the development of the Th2 response to the gastrointestinal nematode parasite Heligmosomoides polygyrus. J Immunol. 2003;170(1):384–93. Epub 2002/12/24. doi: 10.4049/jimmunol.170.1.384 . - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

