Structural characterization of the Sel1-like repeat protein LceB from Legionella pneumophila
- PMID: 38160319
- PMCID: PMC10868440
- DOI: 10.1002/pro.4889
Structural characterization of the Sel1-like repeat protein LceB from Legionella pneumophila
Abstract
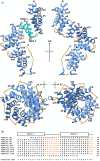
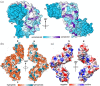
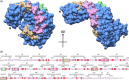
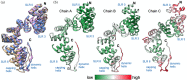
Legionella are freshwater Gram-negative bacteria that in their normal environment infect protozoa. However, this adaptation also allows Legionella to infect human alveolar macrophages and cause pneumonia. Central to Legionella pathogenesis are more than 330 secreted effectors, of which there are nine core effectors that are conserved in all pathogenic species. Despite their importance, the biochemical function of several core effectors remains unclear. To address this, we have taken a structural approach to characterize the core effector of unknown function LceB, or Lpg1356, from Legionella pneumophila. Here, we solve an X-ray crystal structure of LceB using an AlphaFold model for molecular replacement. The experimental structure shows that LceB adopts a Sel1-like repeat (SLR) fold as predicted. However, the crystal structure captured multiple conformations of LceB, all of which differed from the AlphaFold model. A comparison of the predicted model and the experimental models suggests that LceB is highly flexible in solution. Additionally, the molecular analysis of LceB using its close structural homologs reveals sequence and structural motifs of known biochemical function. Specifically, LceB harbors a repeated KAAEQG motif that both stabilizes the SLR fold and is known to participate in protein-protein interactions with eukaryotic host proteins. We also observe that LceB forms several higher-order oligomers in solution. Overall, our results have revealed that LceB has conformational flexibility, self-associates, and contains a molecular surface for binding a target host-cell protein. Additionally, our data provides structural insights into the SLR family of proteins that remain poorly studied.
Keywords: LceB; Legionella pneumophila; Lpg1356; Sel1-like repeat protein; Type IV secretion system; X-ray crystallography; effector.
© 2023 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Similar articles
-
Divergent Evolution of Legionella RCC1 Repeat Effectors Defines the Range of Ran GTPase Cycle Targets.mBio. 2020 Mar 24;11(2):e00405-20. doi: 10.1128/mBio.00405-20. mBio. 2020. PMID: 32209684 Free PMC article.
-
Crystal structure of the Legionella pneumophila Lem10 effector reveals a new member of the HD protein superfamily.Proteins. 2015 Dec;83(12):2319-25. doi: 10.1002/prot.24933. Epub 2015 Oct 14. Proteins. 2015. PMID: 26426142 Free PMC article.
-
Structural and functional study of Legionella pneumophila effector RavA.Protein Sci. 2021 May;30(5):940-955. doi: 10.1002/pro.4057. Epub 2021 Mar 9. Protein Sci. 2021. PMID: 33660322 Free PMC article.
-
The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors.Cell Microbiol. 2009 Oct;11(10):1435-43. doi: 10.1111/j.1462-5822.2009.01351.x. Epub 2009 Jun 26. Cell Microbiol. 2009. PMID: 19563462 Review.
-
[Host-pathogen interaction of Legionella pneumophila].Nihon Saikingaku Zasshi. 2014;69(3):503-11. doi: 10.3412/jsb.69.503. Nihon Saikingaku Zasshi. 2014. PMID: 25186641 Review. Japanese.
References
-
- Allison MJ, Dawson KA, Mayberry WR, Foss JG. Oxalobacter formigenes gen. Nov., sp. nov.: oxalate‐degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol. 1985;141:1–7. - PubMed
-
- Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–423. - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA. In: Struhl K, editor. Short protocols in molecular biology: a compendium of methods from current protocols in molecular biology. 5th ed. New York: John Wiley & Sons Inc.; 2002.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

