A comprehensive evolutionary scenario for the origin and neofunctionalization of the Drosophila speciation gene Odysseus (OdsH)
- PMID: 38156703
- PMCID: PMC10917504
- DOI: 10.1093/g3journal/jkad299
A comprehensive evolutionary scenario for the origin and neofunctionalization of the Drosophila speciation gene Odysseus (OdsH)
Abstract
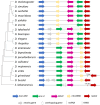
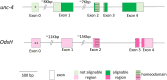
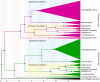
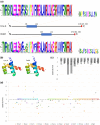
Odysseus (OdsH) was the first speciation gene described in Drosophila related to hybrid sterility in offspring of mating between Drosophila mauritiana and Drosophila simulans. Its origin is attributed to the duplication of the gene unc-4 in the subgenus Sophophora. By using a much larger sample of Drosophilidae species, we showed that contrary to what has been previously proposed, OdsH origin occurred 62 MYA. Evolutionary rates, expression, and transcription factor-binding sites of OdsH evidence that it may have rapidly experienced neofunctionalization in male sexual functions. Furthermore, the analysis of the OdsH peptide allowed the identification of mutations of D. mauritiana that could result in incompatibility in hybrids. In order to find if OdsH could be related to hybrid sterility, beyond Sophophora, we explored the expression of OdsH in Drosophila arizonae and Drosophila mojavensis, a pair of sister species with incomplete reproductive isolation. Our data indicated that OdsH expression is not atypical in their male-sterile hybrids. In conclusion, we have proposed that the origin of OdsH occurred earlier than previously proposed, followed by neofunctionalization. Our results also suggested that its role as a speciation gene might be restricted to D. mauritiana and D. simulans.
Keywords: OdsH expression; unc-4; Drosophilidae; gene duplication; homeodomain; transcription factor.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflicts of interest.
Figures

Similar articles
-
Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species.Science. 2009 Dec 11;326(5959):1538-41. doi: 10.1126/science.1181756. Science. 2009. PMID: 19933102 Free PMC article.
-
Gene duplication and speciation in Drosophila: evidence from the Odysseus locus.Proc Natl Acad Sci U S A. 2004 Aug 17;101(33):12232-5. doi: 10.1073/pnas.0401975101. Epub 2004 Aug 10. Proc Natl Acad Sci U S A. 2004. PMID: 15304653 Free PMC article.
-
Strong purifying selection on the Odysseus gene in two clades of sibling species of the Drosophila montium species subgroup.J Mol Evol. 2006 May;62(5):659-62. doi: 10.1007/s00239-005-0101-8. Epub 2006 Apr 11. J Mol Evol. 2006. PMID: 16612548
-
Speciation genetics: epistasis, conflict and the origin of species.Curr Biol. 2007 Feb 20;17(4):R125-7. doi: 10.1016/j.cub.2006.12.030. Curr Biol. 2007. PMID: 17307044 Review.
-
Hybrid Sterility, Genetic Conflict and Complex Speciation: Lessons From the Drosophila simulans Clade Species.Front Genet. 2021 Jun 23;12:669045. doi: 10.3389/fgene.2021.669045. eCollection 2021. Front Genet. 2021. PMID: 34249091 Free PMC article. Review.
Cited by
-
Selection with two alleles of X-linkage and its application to the fitness component analysis of OdsH in Drosophila.G3 (Bethesda). 2024 Sep 4;14(9):jkae157. doi: 10.1093/g3journal/jkae157. G3 (Bethesda). 2024. PMID: 39001870 Free PMC article.
References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. 2015. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. doi:10.1016/j.softx.2015.06.001. - DOI
-
- Altenhoff AM, Glover NM, Dessimoz C. 2019. Inferring orthology and paralogy. In: Anisimova M, editors. Evolutionary Genomics. New York: Humana. p. 149–175. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
