ALK signaling primes the DNA damage response sensitizing ALK-driven neuroblastoma to therapeutic ATR inhibition
- PMID: 38154064
- PMCID: PMC10769851
- DOI: 10.1073/pnas.2315242121
ALK signaling primes the DNA damage response sensitizing ALK-driven neuroblastoma to therapeutic ATR inhibition
Abstract
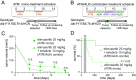
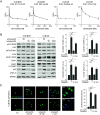
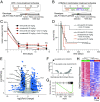
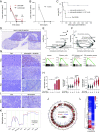
High-risk neuroblastoma (NB) is a significant clinical challenge. MYCN and Anaplastic Lymphoma Kinase (ALK), which are often involved in high-risk NB, lead to increased replication stress in cancer cells, suggesting therapeutic strategies. We previously identified an ATR (ataxia telangiectasia and Rad3-related)/ALK inhibitor (ATRi/ALKi) combination as such a strategy in two independent genetically modified mouse NB models. Here, we identify an underlying molecular mechanism, in which ALK signaling leads to phosphorylation of ATR and CHK1, supporting an effective DNA damage response. The importance of ALK inhibition is supported by mouse data, in which ATRi monotreatment resulted in a robust initial response, but subsequent relapse, in contrast to a 14-d ALKi/ATRi combination treatment that resulted in a robust and sustained response. Finally, we show that the remarkable response to the 14-d combined ATR/ALK inhibition protocol reflects a robust differentiation response, reprogramming tumor cells to a neuronal/Schwann cell lineage identity. Our results identify an ability of ATR inhibition to promote NB differentiation and underscore the importance of further exploring combined ALK/ATR inhibition in NB, particularly in high-risk patient groups with oncogene-induced replication stress.
Keywords: ALK; ATR; elimusertib; lorlatinib; neuroblastoma.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


Similar articles
-
ATR inhibition enables complete tumour regression in ALK-driven NB mouse models.Nat Commun. 2021 Nov 24;12(1):6813. doi: 10.1038/s41467-021-27057-2. Nat Commun. 2021. PMID: 34819497 Free PMC article.
-
The second-generation ALK inhibitor alectinib effectively induces apoptosis in human neuroblastoma cells and inhibits tumor growth in a TH-MYCN transgenic neuroblastoma mouse model.Cancer Lett. 2017 Aug 1;400:61-68. doi: 10.1016/j.canlet.2017.04.022. Epub 2017 Apr 26. Cancer Lett. 2017. PMID: 28455243 Free PMC article.
-
Proliferation and Survival of Embryonic Sympathetic Neuroblasts by MYCN and Activated ALK Signaling.J Neurosci. 2016 Oct 5;36(40):10425-10439. doi: 10.1523/JNEUROSCI.0183-16.2016. J Neurosci. 2016. PMID: 27707976 Free PMC article.
-
Entrectinib and other ALK/TRK inhibitors for the treatment of neuroblastoma.Drug Des Devel Ther. 2018 Oct 23;12:3549-3561. doi: 10.2147/DDDT.S147384. eCollection 2018. Drug Des Devel Ther. 2018. PMID: 30425456 Free PMC article. Review.
-
Targeting anaplastic lymphoma kinase in neuroblastoma.APMIS. 2019 May;127(5):288-302. doi: 10.1111/apm.12940. Epub 2019 Apr 3. APMIS. 2019. PMID: 30803032 Free PMC article. Review.
Cited by
-
Anaplastic Lymphoma Kinase signaling stabilizes SLC3A2 expression via MARCH11 to promote neuroblastoma cell growth.Cell Death Differ. 2024 Jul;31(7):910-923. doi: 10.1038/s41418-024-01319-0. Epub 2024 Jun 10. Cell Death Differ. 2024. PMID: 38858548 Free PMC article.
-
Preclinical exploration of the DNA damage response pathway using the interactive neuroblastoma cell line explorer CLEAN.NAR Cancer. 2024 Jan 11;6(1):zcad062. doi: 10.1093/narcan/zcad062. eCollection 2024 Mar. NAR Cancer. 2024. PMID: 38213997 Free PMC article.
-
Neural crest development and disorders: from patient to model system and back again - the NEUcrest conference.Biol Open. 2024 Jun 15;13(6):bio060530. doi: 10.1242/bio.060530. Epub 2024 Jun 13. Biol Open. 2024. PMID: 38874999 Free PMC article.
References
-
- Matthay K. K., et al. , Neuroblastoma. Nat. Rev. Dis. Primers 2, 16078 (2016). - PubMed
-
- Ladenstein R., et al. , Busulfan and melphalan versus carboplatin, etoposide, and melphalan as high-dose chemotherapy for high-risk neuroblastoma (HR-NBL1/SIOPEN): An international, randomised, multi-arm, open-label, phase 3 trial. Lancet Oncol. 18, 500–514 (2017). - PubMed
-
- De Brouwer S., et al. , Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin. Cancer Res. 16, 4353–4362 (2010). - PubMed
MeSH terms
Substances
Grants and funding
- CAN21/01549/Cancerfonden (Swedish Cancer Society)
- CAN21/1525/Cancerfonden (Swedish Cancer Society)
- 2019-0078/Barncancerfonden (Swedish Childhood Cancer Foundation)
- 2021-0027/Barncancerfonden (Swedish Childhood Cancer Foundation)
- 2019-03914/Vetenskapsrådet (VR)
- 2021-1192/Vetenskapsrådet (VR)
- RB13- 0204/Stiftelsen för Strategisk Forskning (SSF)
- RHP2016/Göran Gustafssons Stiftelser (Göran Gustafsson Foundation)
- 2015.0144/Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)
- FB22-89/Stiftelsen Assar Gabrielssons Fond (AG Fond)
- 2021-0004/Wenner-Gren Stiftelserna (Wenner-Gren Foundations)
- BOF.STG.2019.0073.01/Ghent University Special Research Fund Starting Grant
- V424522N/Research Foundation Flanders
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

