Enhanced Susceptibility to Tomato Chlorosis Virus (ToCV) in Hsp90- and Sgt1-Silenced Plants: Insights from Gene Expression Dynamics
- PMID: 38140611
- PMCID: PMC10747942
- DOI: 10.3390/v15122370
Enhanced Susceptibility to Tomato Chlorosis Virus (ToCV) in Hsp90- and Sgt1-Silenced Plants: Insights from Gene Expression Dynamics
Abstract
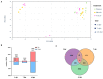
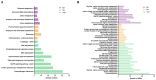
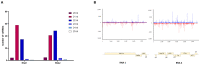
The emerging whitefly-transmitted crinivirus tomato chlorosis virus (ToCV) causes substantial economic losses by inducing yellow leaf disorder in tomato crops. This study explores potential resistance mechanisms by examining early-stage molecular responses to ToCV. A time-course transcriptome analysis compared naïve, mock, and ToCV-infected plants at 2, 7, and 14 days post-infection (dpi). Gene expression changes were most notable at 2 and 14 dpi, likely corresponding to whitefly feeding and viral infection. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses revealed key genes and pathways associated with ToCV infection, including those related to plant immunity, flavonoid and steroid biosynthesis, photosynthesis, and hormone signaling. Additionally, virus-derived small interfering RNAs (vsRNAs) originating from ToCV predominantly came from RNA2 and were 22 nucleotides in length. Furthermore, two genes involved in plant immunity, Hsp90 (heat shock protein 90) and its co-chaperone Sgt1 (suppressor of the G2 allele of Skp1) were targeted through viral-induced gene silencing (VIGS), showing a potential contribution to basal resistance against viral infections since their reduction correlated with increased ToCV accumulation. This study provides insights into tomato plant responses to ToCV, with potential implications for developing effective disease control strategies.
Keywords: Bemisia tabaci; Hsp90; Sgt1; ToCV; basal resistance; tomato.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Transcriptome analysis of the whitefly, Bemisia tabaci MEAM1 during feeding on tomato infected with the crinivirus, Tomato chlorosis virus, identifies a temporal shift in gene expression and differential regulation of novel orphan genes.BMC Genomics. 2017 May 11;18(1):370. doi: 10.1186/s12864-017-3751-1. BMC Genomics. 2017. PMID: 28494755 Free PMC article.
-
Tomato chlorosis virus, an emergent plant virus still expanding its geographical and host ranges.Mol Plant Pathol. 2019 Sep;20(9):1307-1320. doi: 10.1111/mpp.12847. Epub 2019 Jul 2. Mol Plant Pathol. 2019. PMID: 31267719 Free PMC article.
-
Differences in gene expression in whitefly associated with CYSDV-infected and virus-free melon, and comparison with expression in whiteflies fed on ToCV- and TYLCV-infected tomato.BMC Genomics. 2019 Aug 15;20(1):654. doi: 10.1186/s12864-019-5999-0. BMC Genomics. 2019. PMID: 31416422 Free PMC article.
-
Natural Infection Rate of Known Tomato chlorosis virus-Susceptible Hosts and the Influence of the Host Plant on the Virus Relationship With Bemisia tabaci MEAM1.Plant Dis. 2021 May;105(5):1390-1397. doi: 10.1094/PDIS-08-20-1642-RE. Epub 2021 Apr 6. Plant Dis. 2021. PMID: 33107791
-
Epidemiology and genetic diversity of criniviruses associated with tomato yellows disease in Greece.Virus Res. 2014 Jun 24;186:120-9. doi: 10.1016/j.virusres.2013.12.013. Epub 2013 Dec 24. Virus Res. 2014. PMID: 24370865
References
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

