Correlation of MR-Based Metabolomics and Molecular Profiling in the Tumor Microenvironment of Temozolomide-Treated Orthotopic GL261 Glioblastoma in Mice
- PMID: 38139457
- PMCID: PMC10743933
- DOI: 10.3390/ijms242417628
Correlation of MR-Based Metabolomics and Molecular Profiling in the Tumor Microenvironment of Temozolomide-Treated Orthotopic GL261 Glioblastoma in Mice
Abstract
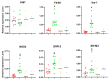
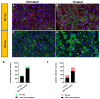
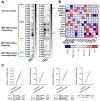
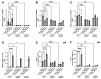
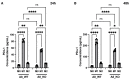
The tumor microenvironment in glioblastoma (GB) is considered to be "cold", i.e., the fraction of cytotoxic T cells, for instance, is low. Instead, macrophages are the major immune cell population in GB, which stem either from tissue response (resident microglia) or recruitment of macrophages from the periphery, thereby undergoing tumor-dependent "imprinting" mechanisms by which macrophages can adapt a tumor-supportive phenotype. In this regard, it is important to describe the nature of macrophages associated with GB, in particular under therapy conditions using the gold standard chemotherapy drug temozolomide (TMZ). Here, we explored the suitability of combining information from in vivo magnetic resonance spectroscopic (MRS) approaches (metabolomics) with in vitro molecular analyses to assess therapy response and characterize macrophage populations in mouse GB using an isogenic GL261 model. For macrophage profiling, expression levels of matrix metalloproteinases (MMPs) and A disintegrin and metalloproteinases (ADAMs) were determined, since their gene products affect macrophage-tumor cell communication by extensive cleavage of immunomodulatory membrane proteins, such as PD-L1. In tumor mice with an overall therapy response, expression of genes encoding the proteases ADAM8, ADAM10, and ADAM17 was increased and might contribute to the immunosuppressive phenotype of GB and immune cells. In tumors responding to therapy, expression levels of ADAM8 were upregulated by TMZ, and higher levels of PD-L1 were correlated significantly. Using a CRISPR/Cas9 knockout of ADAM8 in GL261 cells, we demonstrated that soluble PD-L1 (sPD-L1) is only generated in the presence of ADAM8. Moreover, primary macrophages from WT and ADAM8-deficient mice showed ADAM8-dependent release of sPD-L1, independent of the macrophage polarization state. Since ADAM8 expression is induced in responding tumors and PD-L1 shedding is likely to decrease the anti-tumor activities of T-cells, we conclude that immunotherapy resistance is caused, at least in part, by the increased presence of proteases, such as ADAM8.
Keywords: MR spectroscopic imaging; PD-L1; glioblastoma; macrophages; metalloproteases; shedding; temozolomide; therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Anti-tumour immune response in GL261 glioblastoma generated by Temozolomide Immune-Enhancing Metronomic Schedule monitored with MRSI-based nosological images.NMR Biomed. 2020 Apr;33(4):e4229. doi: 10.1002/nbm.4229. Epub 2020 Jan 11. NMR Biomed. 2020. PMID: 31926117
-
Combination of p38 MAPK inhibitor with PD-L1 antibody effectively prolongs survivals of temozolomide-resistant glioma-bearing mice via reduction of infiltrating glioma-associated macrophages and PD-L1 expression on resident glioma-associated microglia.Brain Tumor Pathol. 2021 Jul;38(3):189-200. doi: 10.1007/s10014-021-00404-3. Epub 2021 Jul 6. Brain Tumor Pathol. 2021. PMID: 34231121
-
The GBM Tumor Microenvironment as a Modulator of Therapy Response: ADAM8 Causes Tumor Infiltration of Tams through HB-EGF/EGFR-Mediated CCL2 Expression and Overcomes TMZ Chemosensitization in Glioblastoma.Cancers (Basel). 2022 Oct 7;14(19):4910. doi: 10.3390/cancers14194910. Cancers (Basel). 2022. PMID: 36230833 Free PMC article.
-
Challenges and potential of PD-1/PD-L1 checkpoint blockade immunotherapy for glioblastoma.J Exp Clin Cancer Res. 2019 Feb 18;38(1):87. doi: 10.1186/s13046-019-1085-3. J Exp Clin Cancer Res. 2019. PMID: 30777100 Free PMC article. Review.
-
A jack of all trades - ADAM8 as a signaling hub in inflammation and cancer.FEBS J. 2024 Sep;291(18):3989-4008. doi: 10.1111/febs.17034. Epub 2023 Dec 22. FEBS J. 2024. PMID: 38097912 Review.
References
MeSH terms
Substances
Grants and funding
- PID2020-113058GB-I00/Centro de Investigación Biomédica en Red-Bioingeniería, Biomateriales y Nanomedicina
- PID2021-128717OB-I00/Centro de Investigación Biomédica en Red-Bioingeniería, Biomateriales y Nanomedicina
- 01KU1915B (PerProGlio)/Federal Ministry of Education and Research (BMBF) Germany
- 11/2023 MR/Universitätsklinikum Gießen und Marburg
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

