ATRX is a predictive marker for endocrinotherapy and chemotherapy resistance in HER2-/HR+ breast cancer through the regulation of the AR, GLI3 and GATA2 transcriptional network
- PMID: 38126976
- PMCID: PMC10781474
- DOI: 10.18632/aging.205327
ATRX is a predictive marker for endocrinotherapy and chemotherapy resistance in HER2-/HR+ breast cancer through the regulation of the AR, GLI3 and GATA2 transcriptional network
Abstract
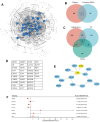
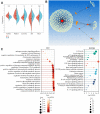
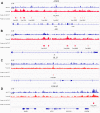
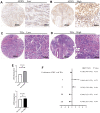
Drug resistance in breast cancer (BC) is a clinical challenge. Exploring the mechanism and identifying a precise predictive biomarker for the drug resistance in BC is critical. Three first-line drug (paclitaxel, doxorubicin and tamoxifen) resistance datasets in BC from GEO were merged to obtain 1,461 differentially expressed genes for weighted correlation network analysis, resulting in identifying ATRX as the hub gene. ATRX is a chromatin remodelling protein, therefore, ATRX-associated transcription factors were explored, thereby identifying the network of AR, GLI3 and GATA2. GO and KEGG analyses revealed immunity, transcriptional regulation and endocrinotherapy/chemotherapy resistance were enriched. Moreover, CIBERSORT revealed immunity regulation was inhibited in the resistance group. ssGSEA showed a significantly lower immune status in the ATRX-Low group compared to the ATRX-High group. Furthermore, the peaks of H3K9me3 ChIP-seq on the four genes were higher in normal tissues than in BC tissues. Notably, the frequency of ATRX mutation was higher than BRCA in BC. Moreover, depressed ATRX revealed worse overall survival and disease-free survival in the human epidermal growth factor receptor 2 (HER2)-/hormone receptor (HR)+ BC. Additionally, depressed ATRX predicted poor results for patients who underwent endocrinotherapy or chemotherapy in the HER2-/HR+ BC subgroup. A nomogram based on ATRX, TILs and ER exhibited a significantly accurate survival prediction ability. Importantly, overexpression of ATRX significantly inhibited the IC50 of the three first-line drugs on MCF-7 cell. Thus, ATRX is an efficient predictive biomarker for endocrinotherapy and chemotherapy resistance in HER2-/HR+ BC and acts by suppressing the AR, GLI3 and GATA2 transcriptional network.
Keywords: ATRX; biomarker; breast cancer; drug resistance; transcription factor.
Conflict of interest statement
Figures







Similar articles
-
Identification of hub genes in AR-induced tamoxifen resistance in breast cancer based on weighted gene co-expression network analysis.Breast Cancer Res Treat. 2023 Jan;197(1):71-82. doi: 10.1007/s10549-022-06788-w. Epub 2022 Nov 5. Breast Cancer Res Treat. 2023. PMID: 36334189
-
Tumour-infiltrating lymphocytes as a prognostic and tamoxifen predictive marker in premenopausal breast cancer: data from a randomised trial with long-term follow-up.Breast Cancer Res. 2020 Dec 23;22(1):140. doi: 10.1186/s13058-020-01364-w. Breast Cancer Res. 2020. PMID: 33357231 Free PMC article. Clinical Trial.
-
AR pathway activity correlates with AR expression in a HER2-dependent manner and serves as a better prognostic factor in breast cancer.Cell Oncol (Dordr). 2020 Apr;43(2):321-333. doi: 10.1007/s13402-019-00492-6. Epub 2020 Jan 13. Cell Oncol (Dordr). 2020. PMID: 31933152
-
Ki67 and lymphocytes in the pretherapeutic core biopsy of primary invasive breast cancer: positive markers of therapy response prediction and superior survival.Horm Mol Biol Clin Investig. 2017 Sep 22;32(2):/j/hmbci.2017.32.issue-2/hmbci-2017-0022/hmbci-2017-0022.xml. doi: 10.1515/hmbci-2017-0022. Horm Mol Biol Clin Investig. 2017. PMID: 28937963 Review.
-
Inhibition of erbB receptor (HER) tyrosine kinases as a strategy to abrogate antiestrogen resistance in human breast cancer.Clin Cancer Res. 2001 Dec;7(12 Suppl):4436s-4442s; discussion 4411s-4412s. Clin Cancer Res. 2001. PMID: 11916237 Review.
References
-
- Dodson A, Parry S, Ibrahim M, Bartlett JM, Pinder S, Dowsett M, Miller K. Breast cancer biomarkers in clinical testing: analysis of a UK national external quality assessment scheme for immunocytochemistry and in situ hybridisation database containing results from 199 300 patients. J Pathol Clin Res. 2018; 4:262–73. 10.1002/cjp2.112 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

