Monovalent metal ion binding promotes the first transesterification reaction in the spliceosome
- PMID: 38123540
- PMCID: PMC10733407
- DOI: 10.1038/s41467-023-44174-2
Monovalent metal ion binding promotes the first transesterification reaction in the spliceosome
Abstract
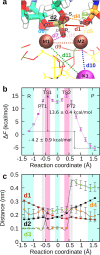
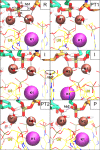
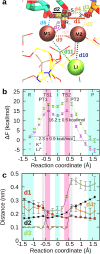
Cleavage and formation of phosphodiester bonds in nucleic acids is accomplished by large cellular machineries composed of both protein and RNA. Long thought to rely on a two-metal-ion mechanism for catalysis, structure comparisons revealed many contain highly spatially conserved second-shell monovalent cations, whose precise function remains elusive. A recent high-resolution structure of the spliceosome, essential for pre-mRNA splicing in eukaryotes, revealed a potassium ion in the active site. Here, we employ biased quantum mechanics/ molecular mechanics molecular dynamics to elucidate the function of this monovalent ion in splicing. We discover that the K+ ion regulates the kinetics and thermodynamics of the first splicing step by rigidifying the active site and stabilizing the substrate in the pre- and post-catalytic state via formation of key hydrogen bonds. Our work supports a direct role for the K+ ion during catalysis and provides a mechanistic hypothesis likely shared by other nucleic acid processing enzymes.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Third Metal Ion Dictates the Catalytic Activity of the Two-Metal-Ion Pre-Ribosomal RNA-Processing Machinery.Angew Chem Int Ed Engl. 2024 Oct 24;63(44):e202405819. doi: 10.1002/anie.202405819. Epub 2024 Sep 17. Angew Chem Int Ed Engl. 2024. PMID: 38994644
-
Second-Shell Basic Residues Expand the Two-Metal-Ion Architecture of DNA and RNA Processing Enzymes.Structure. 2018 Jan 2;26(1):40-50.e2. doi: 10.1016/j.str.2017.11.008. Epub 2017 Dec 7. Structure. 2018. PMID: 29225080 Free PMC article.
-
Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: extending the parallels between the spliceosome and group II introns.RNA. 2000 Feb;6(2):199-205. doi: 10.1017/s1355838200992069. RNA. 2000. PMID: 10688359 Free PMC article.
-
The spliceosome and its metal ions.Met Ions Life Sci. 2011;9:235-51. doi: 10.1039/9781849732512-00235. Met Ions Life Sci. 2011. PMID: 22010274 Review.
-
RNA splicing: group I intron crystal structures reveal the basis of splice site selection and metal ion catalysis.Curr Opin Struct Biol. 2006 Jun;16(3):319-26. doi: 10.1016/j.sbi.2006.04.005. Epub 2006 May 11. Curr Opin Struct Biol. 2006. PMID: 16697179 Review.
Cited by
-
Intron-lariat spliceosomes convert lariats to true circles: implications for intron transposition.bioRxiv [Preprint]. 2024 Mar 27:2024.03.26.586863. doi: 10.1101/2024.03.26.586863. bioRxiv. 2024. Update in: Genes Dev. 2024 May 21;38(7-8):322-335. doi: 10.1101/gad.351764.124. PMID: 38585890 Free PMC article. Updated. Preprint.
-
Intron lariat spliceosomes convert lariats to true circles: implications for intron transposition.Genes Dev. 2024 May 21;38(7-8):322-335. doi: 10.1101/gad.351764.124. Genes Dev. 2024. PMID: 38724209 Free PMC article.
-
Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.Nat Commun. 2024 Jun 19;15(1):4980. doi: 10.1038/s41467-024-48697-0. Nat Commun. 2024. PMID: 38898052 Free PMC article.
-
Stimuli-responsive Zn(ii) complexes showing the structural conversion and on/off switching of catalytic properties.RSC Adv. 2024 Oct 16;14(44):32655-32660. doi: 10.1039/d4ra06058j. eCollection 2024 Oct 9. RSC Adv. 2024. PMID: 39416374 Free PMC article.
References
-
- Sgrignani J, Magistrato A. QM/MM MD simulations on the enzymatic pathway of the human flap endonuclease (hFEN1) elucidating common cleavage pathways to RNase H enzymes. ACS Catal. 2015;5:3864–3875. doi: 10.1021/acscatal.5b00178. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

