High-quality genome of the zoophytophagous stink bug, Nesidiocoris tenuis, informs their food habit adaptation
- PMID: 38113473
- PMCID: PMC10849345
- DOI: 10.1093/g3journal/jkad289
High-quality genome of the zoophytophagous stink bug, Nesidiocoris tenuis, informs their food habit adaptation
Abstract
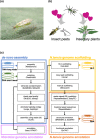
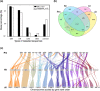
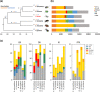
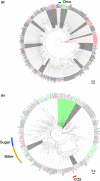
The zoophytophagous stink bug, Nesidiocoris tenuis, is a promising natural enemy of micro-pests such as whiteflies and thrips. This bug possesses both phytophagous and entomophagous food habits, enabling it to obtain nutrition from both plants and insects. This trait allows us to maintain its population density in agricultural fields by introducing insectary plants, even when the pest prey density is extremely low. However, if the bugs' population becomes too dense, they can sometimes damage crop plants. This dual character seems to arise from the food preferences and chemosensation of this predator. To understand the genomic landscape of N. tenuis, we examined the whole genome sequence of a commercially available Japanese strain. We used long-read sequencing and Hi-C analysis to assemble the genome at the chromosomal level. We then conducted a comparative analysis of the genome with previously reported genomes of phytophagous and hematophagous stink bugs to focus on the genetic factors contributing to this species' herbivorous and carnivorous tendencies. Our findings suggest that the gustatory gene set plays a pivotal role in adapting to food habits, making it a promising target for selective breeding. Furthermore, we identified the whole genomes of microorganisms symbiotic with this species through genomic analysis. We believe that our results shed light on the food habit adaptations of N. tenuis and will accelerate breeding efforts based on new breeding techniques for natural enemy insects, including genomics and genome editing.
Keywords: IPM; food habit; natural enemy; zoophytophagous.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflicts of interest.
Figures

Similar articles
-
Interactions between the omnivorous bug Nesidiocoris tenuis (Heteroptera: Miridae) and the tomato pests Helicoverpa armigera (Lepidoptera: Noctuidae) and Phthorimaea absoluta (Lepidoptera: Gelechiidae): predation, phytophagy, and prey preference.J Insect Sci. 2023 Jul 1;23(4):6. doi: 10.1093/jisesa/iead056. J Insect Sci. 2023. PMID: 37428827 Free PMC article.
-
Persistence of the Exotic Mirid Nesidiocoris tenuis (Hemiptera: Miridae) in South Texas.Insects. 2021 Aug 10;12(8):715. doi: 10.3390/insects12080715. Insects. 2021. PMID: 34442281 Free PMC article.
-
Impact of the zoophytophagous plant bug Nesidiocoris tenuis (Heteroptera: Miridae) on tomato yield.J Econ Entomol. 2008 Dec;101(6):1864-70. doi: 10.1603/0022-0493-101.6.1864. J Econ Entomol. 2008. PMID: 19133468
-
Use of zoophytophagous mirid bugs in horticultural crops: Current challenges and future perspectives.Pest Manag Sci. 2021 Jan;77(1):33-42. doi: 10.1002/ps.6043. Epub 2020 Sep 11. Pest Manag Sci. 2021. PMID: 32776672 Review.
-
Dynamics in Pest Status of Phytophagous Stink Bugs in the Neotropics.Neotrop Entomol. 2022 Feb;51(1):18-31. doi: 10.1007/s13744-021-00928-5. Epub 2022 Jan 13. Neotrop Entomol. 2022. PMID: 35028921 Review.
Cited by
-
Chromosome-level genome assembly of the predatory stink bug Arma custos.Sci Data. 2024 Apr 23;11(1):417. doi: 10.1038/s41597-024-03270-8. Sci Data. 2024. PMID: 38654007 Free PMC article.
-
Rickettsia induces strong cytoplasmic incompatibility in a predatory insect.Proc Biol Sci. 2024 Jul;291(2027):20240680. doi: 10.1098/rspb.2024.0680. Epub 2024 Jul 31. Proc Biol Sci. 2024. PMID: 39079670 Free PMC article.
References
-
- Bagci C, Patz S, Huson DH. 2021. Diamond+megan: fast and easy taxonomic and functional analysis of short and long microbiome sequences. Curr Protoc. 1(3):e59. - PubMed
-
- Bailey E, Field L, Rawlings C, King R, Mohareb F, Pak KH, Hughes D, Williamson M, Ganko E, Buer B, et al. 2022. A scaffold-level genome assembly of a minute pirate bug, Orius laevigatus (Hemiptera: Anthocoridae), and a comparative analysis of insecticide resistance-related gene families with hemipteran crop pests. BMC Genomics. 23(1):45. doi:10.1186/s12864-021-08249-y - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
