Dissection of FOXO1-Induced LYPLAL1-DT Impeding Triple-Negative Breast Cancer Progression via Mediating hnRNPK/β-Catenin Complex
- PMID: 38111678
- PMCID: PMC10726293
- DOI: 10.34133/research.0289
Dissection of FOXO1-Induced LYPLAL1-DT Impeding Triple-Negative Breast Cancer Progression via Mediating hnRNPK/β-Catenin Complex
Abstract
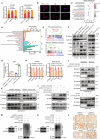
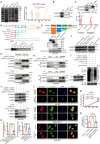
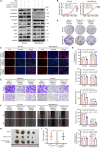
Triple-negative breast cancer (TNBC) is considered as the most hazardous subtype of breast cancer owing to its accelerated progression, enormous metastatic potential, and refractoriness to standard treatments. Long noncoding RNAs (lncRNAs) are extremely intricate in tumorigenesis and cancerous metastasis. Nonetheless, their roles in the initiation and augmentation of TNBC remain elusive. Here, in silico analysis and validation experiments were utilized to analyze the expression pattern of clinically effective lncRNAs in TNBC, among which a protective lncRNA LYPLAL1-DT was essentially curbed in TNBC samples and indicated a favorable prognosis. Gain- and loss-of-function assays elucidated that LYPLAL1-DT considerably attenuated the proliferative and metastatic properties along with epithelial-mesenchymal transition of TNBC cells. Moreover, forkhead box O1 (FOXO1) was validated to modulate the transcription of LYPLAL1-DT. Mechanistically, LYPLAL1-DT impinged on the malignancy of TNBC mainly by restraining the aberrant reactivation of the Wnt/β-catenin signaling pathway, explicitly destabilizing and diminishing β-catenin protein by interacting with heterogeneous nuclear ribonucleoprotein K (hnRNPK) and constricting the formation of the hnRNPK/β-catenin complex. Conclusively, our present research revealed the anti-oncogenic effects of LYPLAL1-DT in TNBC, unraveling the molecular mechanisms of the FOXO1/LYPLAL1-DT/hnRNPK/β-catenin signaling axis, which shed innovative light on the potential curative medicine of TNBC.
Copyright © 2023 Yuhui Tang et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures





Similar articles
-
Role of microRNA/lncRNA Intertwined With the Wnt/β-Catenin Axis in Regulating the Pathogenesis of Triple-Negative Breast Cancer.Front Pharmacol. 2022 Jun 24;13:814971. doi: 10.3389/fphar.2022.814971. eCollection 2022. Front Pharmacol. 2022. PMID: 35814205 Free PMC article. Review.
-
LncRNA LYPLAL1-DT screening from type 2 diabetes with macrovascular complication contributes protective effects on human umbilical vein endothelial cells via regulating the miR-204-5p/SIRT1 axis.Cell Death Discov. 2022 May 4;8(1):245. doi: 10.1038/s41420-022-01019-z. Cell Death Discov. 2022. PMID: 35508613 Free PMC article.
-
The natural compound Jatrophone interferes with Wnt/β-catenin signaling and inhibits proliferation and EMT in human triple-negative breast cancer.PLoS One. 2017 Dec 27;12(12):e0189864. doi: 10.1371/journal.pone.0189864. eCollection 2017. PLoS One. 2017. PMID: 29281678 Free PMC article.
-
FOXO1-regulated lncRNA LINC01197 inhibits pancreatic adenocarcinoma cell proliferation by restraining Wnt/β-catenin signaling.J Exp Clin Cancer Res. 2019 Apr 26;38(1):179. doi: 10.1186/s13046-019-1174-3. J Exp Clin Cancer Res. 2019. PMID: 31027497 Free PMC article.
-
Countering Triple Negative Breast Cancer via Impeding Wnt/β-Catenin Signaling, a Phytotherapeutic Approach.Plants (Basel). 2022 Aug 24;11(17):2191. doi: 10.3390/plants11172191. Plants (Basel). 2022. PMID: 36079579 Free PMC article. Review.
Cited by
-
MEIS2 suppresses breast cancer development by downregulating IL10.Cancer Rep (Hoboken). 2024 May;7(5):e2064. doi: 10.1002/cnr2.2064. Cancer Rep (Hoboken). 2024. PMID: 38711262 Free PMC article.
-
NDR1/FBXO11 promotes phosphorylation-mediated ubiquitination of β-catenin to suppress metastasis in prostate cancer.Int J Biol Sci. 2024 Sep 16;20(12):4957-4977. doi: 10.7150/ijbs.98907. eCollection 2024. Int J Biol Sci. 2024. PMID: 39309441 Free PMC article.
-
Crosstalk between lncRNAs and Wnt/β-catenin signaling pathways in lung cancers: From cancer progression to therapeutic response.Noncoding RNA Res. 2024 Mar 14;9(3):667-677. doi: 10.1016/j.ncrna.2024.02.013. eCollection 2024 Sep. Noncoding RNA Res. 2024. PMID: 38577016 Free PMC article. Review.
-
Long non-coding RNAs: regulators of autophagy and potential biomarkers in therapy resistance and urological cancers.Front Pharmacol. 2024 Oct 24;15:1442227. doi: 10.3389/fphar.2024.1442227. eCollection 2024. Front Pharmacol. 2024. PMID: 39512820 Free PMC article. Review.
-
Prognosis comparison between hepatocellular carcinoma patients with microvascular invasion who received hepatectomy alone and those who underwent early PA-TACE: a retrospective cohort study.J Gastrointest Oncol. 2024 Jun 30;15(3):1112-1121. doi: 10.21037/jgo-24-282. Epub 2024 Jun 17. J Gastrointest Oncol. 2024. PMID: 38989441 Free PMC article.
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. - PubMed
-
- Wu SY, Wang H, Shao ZM, Jiang YZ. Triple-negative breast cancer: New treatment strategies in the era of precision medicine. Sci China Life Sci. 2021;64(3):372–388. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

