Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2
- PMID: 38109549
- PMCID: PMC10756187
- DOI: 10.1073/pnas.2314193120
Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2
Abstract
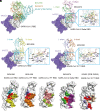
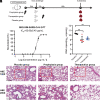
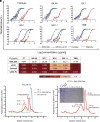
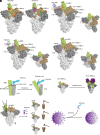
Currently, monoclonal antibodies (MAbs) targeting the SARS-CoV-2 receptor binding domain (RBD) of spike (S) protein are classified into seven classes based on their binding epitopes. However, most of these antibodies are seriously impaired by SARS-CoV-2 Omicron and its subvariants, especially the recent BQ.1.1, XBB and its derivatives. Identification of broadly neutralizing MAbs against currently circulating variants is imperative. In this study, we identified a "breathing" cryptic epitope in the S protein, named as RBD-8. Two human MAbs, BIOLS56 and IMCAS74, were isolated recognizing this epitope with broad neutralization abilities against tested sarbecoviruses, including SARS-CoV, pangolin-origin coronaviruses, and all the SARS-CoV-2 variants tested (Omicron BA.4/BA.5, BQ.1.1, and XBB subvariants). Searching through the literature, some more RBD-8 MAbs were defined. More importantly, BIOLS56 rescues the immune-evaded antibody, RBD-5 MAb IMCAS-L4.65, by making a bispecific MAb, to neutralize BQ.1 and BQ.1.1, thereby producing an MAb to cover all the currently circulating Omicron subvariants. Structural analysis reveals that the neutralization effect of RBD-8 antibodies depends on the extent of epitope exposure, which is affected by the angle of antibody binding and the number of up-RBDs induced by angiotensin-converting enzyme 2 binding. This cryptic epitope which recognizes non- receptor binding motif (non-RBM) provides guidance for the development of universal therapeutic antibodies and vaccines against COVID-19.
Keywords: Omicron BA.4/BA.5/BQ.1.1/XBB subvariants; RBD-8; SARS-CoV-2; cryptic epitope; neutralizing antibody.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



Similar articles
-
A bispecific antibody exhibits broad neutralization against SARS-CoV-2 Omicron variants XBB.1.16, BQ.1.1 and sarbecoviruses.Nat Commun. 2024 Jun 15;15(1):5127. doi: 10.1038/s41467-024-49096-1. Nat Commun. 2024. PMID: 38879565 Free PMC article.
-
A pan-sarbecovirus vaccine based on RBD of SARS-CoV-2 original strain elicits potent neutralizing antibodies against XBB in non-human primates.Proc Natl Acad Sci U S A. 2023 Mar 14;120(11):e2221713120. doi: 10.1073/pnas.2221713120. Epub 2023 Mar 10. Proc Natl Acad Sci U S A. 2023. PMID: 36897979 Free PMC article.
-
An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.Cell Rep Med. 2023 Apr 18;4(4):100991. doi: 10.1016/j.xcrm.2023.100991. Epub 2023 Mar 21. Cell Rep Med. 2023. PMID: 37019110 Free PMC article.
-
Immune evasion of neutralizing antibodies by SARS-CoV-2 Omicron.Cytokine Growth Factor Rev. 2023 Apr;70:13-25. doi: 10.1016/j.cytogfr.2023.03.001. Epub 2023 Mar 5. Cytokine Growth Factor Rev. 2023. PMID: 36948931 Free PMC article. Review.
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
Cited by
-
The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.Virol Sin. 2024 Aug;39(4):609-618. doi: 10.1016/j.virs.2024.06.004. Epub 2024 Jun 10. Virol Sin. 2024. PMID: 38866203 Free PMC article.
-
Facilitating and restraining virus infection using cell-attachable soluble viral receptors.Proc Natl Acad Sci U S A. 2024 Nov 5;121(45):e2414583121. doi: 10.1073/pnas.2414583121. Epub 2024 Oct 31. Proc Natl Acad Sci U S A. 2024. PMID: 39480852 Free PMC article.
-
Broadening sarbecovirus neutralization with bispecific antibodies combining distinct conserved targets on the receptor binding domain.Hum Vaccin Immunother. 2024 Dec 31;20(1):2388344. doi: 10.1080/21645515.2024.2388344. Epub 2024 Aug 20. Hum Vaccin Immunother. 2024. PMID: 39165108 Free PMC article.
References
-
- Wang P., et al. , Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021). - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

