This is a preprint.
RAPIDASH: A tag-free enrichment of ribosome-associated proteins reveals compositional dynamics in embryonic tissues and stimulated macrophages
- PMID: 38106052
- PMCID: PMC10723405
- DOI: 10.1101/2023.12.07.570613
RAPIDASH: A tag-free enrichment of ribosome-associated proteins reveals compositional dynamics in embryonic tissues and stimulated macrophages
Update in
-
RAPIDASH: Tag-free enrichment of ribosome-associated proteins reveals composition dynamics in embryonic tissue, cancer cells, and macrophages.Mol Cell. 2024 Sep 19;84(18):3545-3563.e25. doi: 10.1016/j.molcel.2024.08.023. Epub 2024 Sep 10. Mol Cell. 2024. PMID: 39260367
Abstract
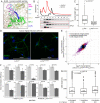
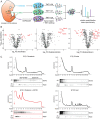
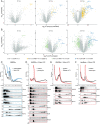
Ribosomes are emerging as direct regulators of gene expression, with ribosome-associated proteins (RAPs) allowing ribosomes to modulate translational control. However, a lack of technologies to enrich RAPs across many sample types has prevented systematic analysis of RAP number, dynamics, and functions. Here, we have developed a label-free methodology called RAPIDASH to enrich ribosomes and RAPs from any sample. We applied RAPIDASH to mouse embryonic tissues and identified hundreds of potential RAPs, including DHX30 and LLPH, two forebrain RAPs important for neurodevelopment. We identified a critical role of LLPH in neural development that is linked to the translation of genes with long coding sequences. Finally, we characterized ribosome composition remodeling during immune activation and observed extensive changes post-stimulation. RAPIDASH has therefore enabled the discovery of RAPs ranging from those with neuroregulatory functions to those activated by immune stimuli, thereby providing critical insights into how ribosomes are remodeled.
Keywords: embryonic development; macrophages; proteomics; ribosome; ribosome heterogeneity; ribosome-associated proteins; translational control.
Conflict of interest statement
Declaration of Interest The authors declare no competing interests.
Figures


Similar articles
-
RAPIDASH: Tag-free enrichment of ribosome-associated proteins reveals composition dynamics in embryonic tissue, cancer cells, and macrophages.Mol Cell. 2024 Sep 19;84(18):3545-3563.e25. doi: 10.1016/j.molcel.2024.08.023. Epub 2024 Sep 10. Mol Cell. 2024. PMID: 39260367
-
The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity.Cell. 2017 Jun 1;169(6):1051-1065.e18. doi: 10.1016/j.cell.2017.05.022. Cell. 2017. PMID: 28575669 Free PMC article.
-
Ribosome heterogeneity in stem cells and development.J Cell Biol. 2020 Jun 1;219(6):e202001108. doi: 10.1083/jcb.202001108. J Cell Biol. 2020. PMID: 32330234 Free PMC article. Review.
-
Proteomic analysis of nascent polypeptide chains that potentially induce translational pausing during elongation.Biosci Biotechnol Biochem. 2022 Aug 24;86(9):1262-1269. doi: 10.1093/bbb/zbac097. Biosci Biotechnol Biochem. 2022. PMID: 35749475
-
Ribosomal heterogeneity - A new inroad for pharmacological innovation.Biochem Pharmacol. 2020 May;175:113874. doi: 10.1016/j.bcp.2020.113874. Epub 2020 Feb 24. Biochem Pharmacol. 2020. PMID: 32105657 Review.
References
-
- Genuth N.R., Shi Z., Kunimoto K., Hung V., Xu A.F., Kerr C.H., Tiu G.C., Oses-Prieto J.A., Salomon-Shulman R.E.A., Axelrod J.D., et al. (2022). A stem cell roadmap of ribosome heterogeneity reveals a function for RPL10A in mesoderm production. Nat Commun 13, 5491. 10.1038/s41467-022-33263-3. - DOI - PMC - PubMed
-
- Bartsch D., Kalamkar K., Ahuja G., Lackmann J.-W., Hescheler J., Weber T., Bazzi H., Clamer M., Mendjan S., Papantonis A., et al. (2023). mRNA translational specialization by RBPMS presets the competence for cardiac commitment in hESCs. Science Advances 9, eade1792. 10.1126/sciadv.ade1792. - DOI - PMC - PubMed
-
- Ricciardi S., Manfrini N., Alfieri R., Calamita P., Crosti M.C., Gallo S., Müller R., Pagani M., Abrignani S., and Biffo S. (2018). The Translational Machinery of Human CD4+ T Cells Is Poised for Activation and Controls the Switch from Quiescence to Metabolic Remodeling. Cell Metabolism 28, 895–906.e5. 10.1016/j.cmet.2018.08.009. - DOI - PMC - PubMed
-
- Koppers M., Cagnetta R., Shigeoka T., Wunderlich L.C., Vallejo-Ramirez P., Qiaojin Lin J., Zhao S., Jakobs M.A., Dwivedy A., Minett M.S., et al. (2019). Receptor-specific interactome as a hub for rapid cue-induced selective translation in axons. eLife 8, e48718. 10.7554/eLife.48718. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
