Identification of hub genes and their expression profiling for predicting buffalo (Bubalus bubalis) semen quality and fertility
- PMID: 38092793
- PMCID: PMC10719284
- DOI: 10.1038/s41598-023-48925-5
Identification of hub genes and their expression profiling for predicting buffalo (Bubalus bubalis) semen quality and fertility
Abstract
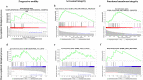
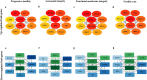
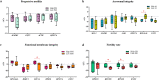
Sperm transcriptomics provide insights into subtle differences in sperm fertilization competence. For predicting the success of complex traits like male fertility, identification of hub genes involved in various sperm functions are essential. The bulls from the transcriptome profiled samples (n = 21), were grouped into good and poor progressive motility (PM), acrosome integrity (AI), functional membrane integrity (FMI) and fertility rate (FR) groups. The up-regulated genes identified in each group were 87, 470, 1715 and 36, respectively. Gene networks were constructed using up- and down-regulated genes from each group. The top clusters from the upregulated gene networks of the PM, AI, FMI and FR groups were involved in tyrosine kinase (FDR = 1.61E-11), apoptosis (FDR = 1.65E-8), translation (FDR = 2.2E-16) and ribosomal pathway (FDR = 1.98E-21), respectively. From the clusters, the hub genes were identified and validated in a fresh set of semen samples (n = 12) using RT-qPCR. Importantly, the genes (fold change) RPL36AL (14.99) in AI, EIF5A (54.32) in FMI, and RPLP0 (8.55) and RPS28 (13.42) in FR were significantly (p < 0.05) up-regulated. The study suggests that the expression levels of MAPK3 (PM), RPL36AL + RPS27A or RPL36AL + EXT2 (AI), RPL36AL or RPS27A (FMI) and RPS18 + RPS28 (FR) are potential markers for diagnosing the semen quality and fertility status of bulls which can be used for the breeding program.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Sulforaphane inclusion in a freezing medium augments post-thaw motility, functional and biochemical features, and fertility potential of buffalo (Bubalus bubalis) spermatozoa.Res Vet Sci. 2023 May;158:196-202. doi: 10.1016/j.rvsc.2023.03.020. Epub 2023 Apr 2. Res Vet Sci. 2023. PMID: 37030095
-
Cryostress induces fragmentation and alters the abundance of sperm transcripts associated with fertilizing competence and reproductive processes in buffalo.Cell Tissue Res. 2023 Jul;393(1):181-199. doi: 10.1007/s00441-023-03764-8. Epub 2023 Apr 20. Cell Tissue Res. 2023. PMID: 37079096
-
Supplementation of l-tryptophan (an aromatic amino acid) in tris citric acid extender enhances post-thaw progressive motility, plasmalemma, mitochondrial membrane potential, acrosome, and DNA integrities, and in vivo fertility rate of buffalo (Bubalus bubalis) bull spermatozoa.Cryobiology. 2020 Feb 1;92:117-123. doi: 10.1016/j.cryobiol.2019.11.044. Epub 2019 Nov 26. Cryobiology. 2020. PMID: 31783000
-
Use of post-thaw semen quality parameters to predict fertility of water buffalo (Bubalus bubalis) bull during peak breeding season.Andrologia. 2017 May;49(4). doi: 10.1111/and.12639. Epub 2016 Jul 12. Andrologia. 2017. PMID: 27401143
-
Current status of sperm functional genomics and its diagnostic potential of fertility in bovine (Bos taurus).Syst Biol Reprod Med. 2018 Dec;64(6):484-501. doi: 10.1080/19396368.2018.1444816. Epub 2018 Mar 14. Syst Biol Reprod Med. 2018. PMID: 29537884 Review.
Cited by
-
Determinant genetic markers of semen quality in livestock.Front Endocrinol (Lausanne). 2024 Oct 4;15:1456305. doi: 10.3389/fendo.2024.1456305. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39429738 Free PMC article. Review.
References
-
- Somashekar L, Selvaraju S, Parthipan S, Patil SK, Binsila BK, Venkataswamy MM, Karthik Bhat S, Ravindra JP. Comparative sperm protein profiling in bulls differing in fertility and identification of phosphatidylethanolamine-binding protein 4, a potential fertility marker. Andrology. 2017;5:1032–1051. doi: 10.1111/andr.12404. - DOI - PubMed
-
- Mathur PP, Francispillai M, Vaithinathan S, Agarwal A. NF-κB in male reproduction: A boon or a bane ? Open Reprod. Sci. J. 2011;3:85–91. doi: 10.2174/1874255601103010085. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

