An ncRNA transcriptomics-based approach to design siRNA molecules against SARS-CoV-2 double membrane vesicle formation and accessory genes
- PMID: 38087193
- PMCID: PMC10718025
- DOI: 10.1186/s12879-023-08870-0
An ncRNA transcriptomics-based approach to design siRNA molecules against SARS-CoV-2 double membrane vesicle formation and accessory genes
Abstract
Background: The corona virus SARS-CoV-2 is the causative agent of recent most global pandemic. Its genome encodes various proteins categorized as non-structural, accessory, and structural proteins. The non-structural proteins, NSP1-16, are located within the ORF1ab. The NSP3, 4, and 6 together are involved in formation of double membrane vesicle (DMV) in host Golgi apparatus. These vesicles provide anchorage to viral replicative complexes, thus assist replication inside the host cell. While the accessory genes coded by ORFs 3a, 3b, 6, 7a, 7b, 8a, 8b, 9b, 9c, and 10 contribute in cell entry, immunoevasion, and pathological progression.
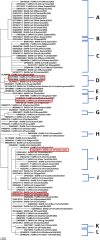
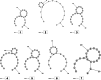
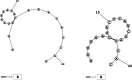
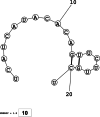
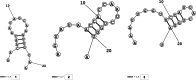
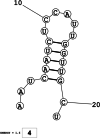
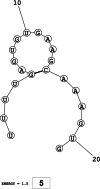
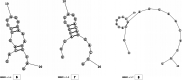
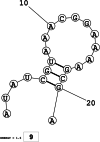
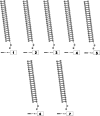
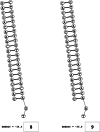
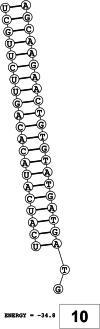
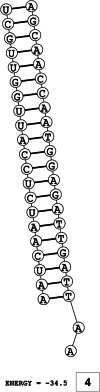
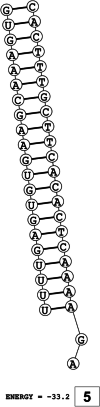
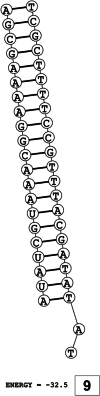
Methods: This in silico study is focused on designing sequence specific siRNA molecules as a tool for silencing the non-structural and accessory genes of the virus. The gene sequences of NSP3, 4, and 6 along with ORF3a, 6, 7a, 8, and 10 were retrieved for conservation, phylogenetic, and sequence logo analyses. siRNA candidates were predicted using siDirect 2.0 targeting these genes. The GC content, melting temperatures, and various validation scores were calculated. Secondary structures of the guide strands and siRNA-target duplexes were predicted. Finally, tertiary structures were predicted and subjected to structural validations.
Results: This study revealed that NSP3, 4, and 6 and accessory genes ORF3a, 6, 7a, 8, and 10 have high levels of conservation across globally circulating SARS-CoV-2 strains. A total of 71 siRNA molecules were predicted against the selected genes. Following rigorous screening including binary validations and minimum free energies, final siRNAs with high therapeutic potential were identified, including 7, 2, and 1 against NSP3, NSP4, and NSP6, as well as 3, 1, 2, and 1 targeting ORF3a, ORF7a, ORF8, and ORF10, respectively.
Conclusion: Our novel in silico pipeline integrates effective methods from previous studies to predict and validate siRNA molecules, having the potential to inhibit viral replication pathway in vitro. In total, this study identified 17 highly specific siRNA molecules targeting NSP3, 4, and 6 and accessory genes ORF3a, 7a, 8, and 10 of SARS-CoV-2, which might be used as an additional antiviral treatment option especially in the cases of life-threatening urgencies.
Keywords: Accessory genes; Gene silencing; NSPs; Non-structural genes; Open Reading frame ORF; SARS-CoV-2; ncRNA; siRNA.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







Similar articles
-
Mapping the Nonstructural Transmembrane Proteins of Severe Acute Respiratory Syndrome Coronavirus 2.J Comput Biol. 2021 Sep;28(9):909-921. doi: 10.1089/cmb.2020.0627. Epub 2021 Jun 28. J Comput Biol. 2021. PMID: 34182794 Free PMC article.
-
A computational approach to design potential siRNA molecules as a prospective tool for silencing nucleocapsid phosphoprotein and surface glycoprotein gene of SARS-CoV-2.Genomics. 2021 Jan;113(1 Pt 1):331-343. doi: 10.1016/j.ygeno.2020.12.021. Epub 2020 Dec 13. Genomics. 2021. PMID: 33321203 Free PMC article.
-
Oxysterole-binding protein targeted by SARS-CoV-2 viral proteins regulates coronavirus replication.Front Cell Infect Microbiol. 2024 Jul 25;14:1383917. doi: 10.3389/fcimb.2024.1383917. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39119292 Free PMC article.
-
SARS-CoV-2 Proteins: Are They Useful as Targets for COVID-19 Drugs and Vaccines?Curr Mol Med. 2022;22(1):50-66. doi: 10.2174/1566524021666210223143243. Curr Mol Med. 2022. PMID: 33622224 Review.
-
The role of SARS-CoV-2 accessory proteins in immune evasion.Biomed Pharmacother. 2022 Dec;156:113889. doi: 10.1016/j.biopha.2022.113889. Epub 2022 Oct 17. Biomed Pharmacother. 2022. PMID: 36265309 Free PMC article. Review.
References
-
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard [Internet]. Geneva, Switzerland [updated 2023 17; cited 2023 May 17]. Available from: https://covid19.who.int/
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

