Isolation and detection of DNA-protein crosslinks in mammalian cells
- PMID: 38084926
- PMCID: PMC10810220
- DOI: 10.1093/nar/gkad1178
Isolation and detection of DNA-protein crosslinks in mammalian cells
Abstract
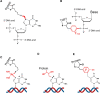
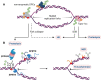
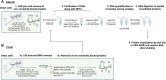
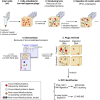
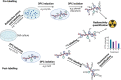
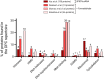
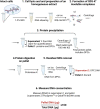
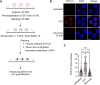
DNA-protein crosslinks (DPCs) are toxic DNA lesions wherein a protein is covalently attached to DNA. If not rapidly repaired, DPCs create obstacles that disturb DNA replication, transcription and DNA damage repair, ultimately leading to genome instability. The persistence of DPCs is associated with premature ageing, cancer and neurodegeneration. In mammalian cells, the repair of DPCs mainly relies on the proteolytic activities of SPRTN and the 26S proteasome, complemented by other enzymes including TDP1/2 and the MRN complex, and many of the activities involved are essential, restricting genetic approaches. For many years, the study of DPC repair in mammalian cells was hindered by the lack of standardised assays, most notably assays that reliably quantified the proteins or proteolytic fragments covalently bound to DNA. Recent interest in the field has spurred the development of several biochemical methods for DPC analysis. Here, we critically analyse the latest techniques for DPC isolation and the benefits and drawbacks of each. We aim to assist researchers in selecting the most suitable isolation method for their experimental requirements and questions, and to facilitate the comparison of results across different laboratories using different approaches.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Tyrosyl-DNA phosphodiesterase 1 (TDP1) and SPRTN protease repair histone 3 and topoisomerase 1 DNA-protein crosslinks in vivo.Open Biol. 2023 Oct;13(10):230113. doi: 10.1098/rsob.230113. Epub 2023 Oct 4. Open Biol. 2023. PMID: 37788708 Free PMC article.
-
The protease SPRTN and SUMOylation coordinate DNA-protein crosslink repair to prevent genome instability.Cell Rep. 2021 Dec 7;37(10):110080. doi: 10.1016/j.celrep.2021.110080. Cell Rep. 2021. PMID: 34879279 Free PMC article.
-
SPRTN patient variants cause global-genome DNA-protein crosslink repair defects.Nat Commun. 2023 Jan 21;14(1):352. doi: 10.1038/s41467-023-35988-1. Nat Commun. 2023. PMID: 36681662 Free PMC article.
-
Formation and repair of DNA-protein crosslink damage.Sci China Life Sci. 2017 Oct;60(10):1065-1076. doi: 10.1007/s11427-017-9183-4. Epub 2017 Oct 30. Sci China Life Sci. 2017. PMID: 29098631 Free PMC article. Review.
-
DNA-Protein Crosslinks and Their Resolution.Annu Rev Biochem. 2022 Jun 21;91:157-181. doi: 10.1146/annurev-biochem-032620-105820. Epub 2022 Mar 18. Annu Rev Biochem. 2022. PMID: 35303790 Review.
Cited by
-
Quantification of Intracellular DNA-Protein Cross-Links with N7-Methyl-2'-Deoxyguanosine and Their Contribution to Cytotoxicity.Chem Res Toxicol. 2024 May 20;37(5):814-823. doi: 10.1021/acs.chemrestox.4c00076. Epub 2024 Apr 23. Chem Res Toxicol. 2024. PMID: 38652696
References
-
- Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001; 70:369–413. - PubMed
-
- Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006; 6:789–802. - PubMed
-
- Weickert P., Stingele J.. DNA–protein crosslinks and their resolution. Annu. Rev. Biochem. 2022; 91:157–181. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

