Specifications of the ACMG/AMP Variant Classification Guidelines for Germline DICER1 Variant Curation
- PMID: 38084291
- PMCID: PMC10713350
- DOI: 10.1155/2023/9537832
Specifications of the ACMG/AMP Variant Classification Guidelines for Germline DICER1 Variant Curation
Abstract
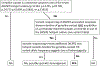
Germline pathogenic variants in DICER1 predispose individuals to develop a variety of benign and malignant tumors. Accurate variant curation and classification is essential for reliable diagnosis of DICER1-related tumor predisposition and identification of individuals who may benefit from surveillance. Since 2015, most labs have followed the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) sequence variant classification guidelines for DICER1 germline variant curation. However, these general guidelines lack gene-specific nuances and leave room for subjectivity. Consequently, a group of DICER1 experts joined ClinGen to form the DICER1 and miRNA-Processing Genes Variant Curation Expert Panel (VCEP), to create DICER1- specific ACMG/AMP guidelines for germline variant curation. The VCEP followed the FDA-approved ClinGen protocol for adapting and piloting these guidelines. A diverse set of 40 DICER1 variants were selected for piloting, including 14 known Pathogenic/Likely Pathogenic (P/LP) variants, 12 known Benign/Likely Benign (B/LB) variants, and 14 variants classified as variants of uncertain significance (VUS) or with conflicting interpretations in ClinVar. Clinically meaningful classifications (i.e., P, LP, LB, or B) were achieved for 82.5% (33/40) of the pilot variants, with 100% concordance among the known P/LP and known B/LB variants. Half of the VUS or conflicting variants were resolved with four variants classified as LB and three as LP. These results demonstrate that the DICER1-specific guidelines for germline variant curation effectively classify known pathogenic and benign variants while reducing the frequency of uncertain classifications. Individuals and labs curating DICER1 variants should consider adopting this classification framework to encourage consistency and improve objectivity.
Keywords: ClinGen; ClinVar; DICER1; cancer predisposition; germline pathogenic variants; pediatric cancer; variant curation.
Conflict of interest statement
The following authors work for laboratories that offer fee-for-service testing of DICER1: MJA, ECC, HCC, SBC, SH, JM, JLM, NNY. The following authors have made substantial contributions to the DICER1 gene: disease literature: DRS, XB-D, KSC, WDF, SG, KAS, MKW. NSA-H is an employee and equity holder of 23andMe; serves as a scientific advisory board member for Allelica; received personal fees from Genentech, Allelica, and 23andMe; received research funding from Akcea; and was previously employed by Regeneron Pharmaceuticals.
Figures
Similar articles
-
Disease-specific ACMG/AMP guidelines improve sequence variant interpretation for hearing loss.Genet Med. 2021 Nov;23(11):2208-2212. doi: 10.1038/s41436-021-01254-2. Epub 2021 Jul 6. Genet Med. 2021. PMID: 34230634 Free PMC article.
-
Specifications of the ACMG/AMP variant curation guidelines for the analysis of germline ATM sequence variants.Am J Hum Genet. 2024 Sep 17:S0002-9297(24)00332-X. doi: 10.1016/j.ajhg.2024.08.022. Online ahead of print. Am J Hum Genet. 2024. PMID: 39317201
-
Specifications of the ACMG/AMP variant curation guidelines for the analysis of germline CDH1 sequence variants.Hum Mutat. 2018 Nov;39(11):1553-1568. doi: 10.1002/humu.23650. Hum Mutat. 2018. PMID: 30311375 Free PMC article.
-
Evaluation of the pathogenic potential of germline DDX41 variants in hematopoietic neoplasms using the ACMG/AMP guidelines.Int J Hematol. 2024 May;119(5):552-563. doi: 10.1007/s12185-024-03728-w. Epub 2024 Mar 16. Int J Hematol. 2024. PMID: 38492200 Review.
-
DICER1 and Associated Conditions: Identification of At-risk Individuals and Recommended Surveillance Strategies.Clin Cancer Res. 2018 May 15;24(10):2251-2261. doi: 10.1158/1078-0432.CCR-17-3089. Epub 2018 Jan 17. Clin Cancer Res. 2018. PMID: 29343557 Free PMC article. Review.
Cited by
-
Co-observation of germline pathogenic variants in breast cancer predisposition genes: Results from analysis of the BRIDGES sequencing dataset.Am J Hum Genet. 2024 Sep 5;111(9):2059-2069. doi: 10.1016/j.ajhg.2024.07.004. Epub 2024 Aug 2. Am J Hum Genet. 2024. PMID: 39096911
-
A genome-first approach to characterize DICER1 pathogenic variant prevalence, penetrance and cancer, thyroid, and other phenotypes in 2 population-scale cohorts.Genet Med Open. 2024;2:101846. doi: 10.1016/j.gimo.2024.101846. Epub 2024 Apr 11. Genet Med Open. 2024. PMID: 39070603
-
Intronic Germline DICER1 Variants in Patients With Sertoli-Leydig Cell Tumor.JCO Precis Oncol. 2023 Sep;7:e2300189. doi: 10.1200/PO.23.00189. JCO Precis Oncol. 2023. PMID: 37883719 Free PMC article.
-
Specifications of the ACMG/AMP guidelines for ACADVL variant interpretation.Mol Genet Metab. 2023 Nov;140(3):107668. doi: 10.1016/j.ymgme.2023.107668. Epub 2023 Jul 26. Mol Genet Metab. 2023. PMID: 37549443 Free PMC article.
-
Reclassification of two germline DICER1 splicing variants leads to DICER1 syndrome diagnosis.Fam Cancer. 2023 Oct;22(4):487-493. doi: 10.1007/s10689-023-00336-1. Epub 2023 May 30. Fam Cancer. 2023. PMID: 37248399 Free PMC article.
References
-
- Altaraihi M, Hansen TVO, Santoni-Rugiu E, Rossing M, Rasmussen Å K, Gerdes AM, & Wadt K (2021). Prevalence of Pathogenic Germline DICER1 Variants in Young Individuals Thyroidectomised Due to Goitre - A National Danish Cohort. Front Endocrinol (Lausanne), 12, 727970. 10.3389/fendo.2021.727970 - DOI - PMC - PubMed
-
- Apellaniz-Ruiz M, Segni M, Kettwig M, Glüer S, Pelletier D, Nguyen VH, Wagener R, López C, Muchantef K, Bouron-Dal Soglio D, Sabbaghian N, Wu MK, Zannella S, Fabian MR, Siebert R, Menke J, Priest JR, & Foulkes WD (2019). Mesenchymal Hamartoma of the Liver and DICER1 Syndrome. N Engl J Med, 380(19), 1834–1842. 10.1056/NEJMoa1812169 - DOI - PubMed
-
- Bakhuizen JJ, Hanson H, van der Tuin K, Lalloo F, Tischkowitz M, Wadt K, & Jongmans MCJ (2021). Surveillance recommendations for DICER1 pathogenic variant carriers: a report from the SIOPE Host Genome Working Group and CanGene-CanVar Clinical Guideline Working Group. Fam Cancer, 20(4), 337–348. 10.1007/s10689-021-00264-y - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials