E. coli RecB Nuclease Domain Regulates RecBCD Helicase Activity but not Single Stranded DNA Translocase Activity
- PMID: 38081382
- PMCID: PMC11131135
- DOI: 10.1016/j.jmb.2023.168381
E. coli RecB Nuclease Domain Regulates RecBCD Helicase Activity but not Single Stranded DNA Translocase Activity
Abstract
Much is still unknown about the mechanisms by which helicases unwind duplex DNA. Whereas structure-based models describe DNA unwinding as occurring by the ATPase motors mechanically pulling the DNA duplex across a wedge domain in the helicase, biochemical data show that processive DNA unwinding by E. coli RecBCD helicase can occur in the absence of ssDNA translocation by the canonical RecB and RecD motors. Here we show that DNA unwinding is not a simple consequence of ssDNA translocation by the motors. Using stopped-flow fluorescence approaches, we show that a RecB nuclease domain deletion variant (RecBΔNucCD) unwinds dsDNA at significantly slower rates than RecBCD, while the ssDNA translocation rate is unaffected. This effect is primarily due to the absence of the nuclease domain since a nuclease-dead mutant (RecBD1080ACD), which retains the nuclease domain, showed no change in ssDNA translocation or dsDNA unwinding rates relative to RecBCD on short DNA substrates (≤60 base pairs). Hence, ssDNA translocation is not rate-limiting for DNA unwinding. RecBΔNucCD also initiates unwinding much slower than RecBCD from a blunt-ended DNA. RecBΔNucCD also unwinds DNA ∼two-fold slower than RecBCD on long DNA (∼20 kilo base pair) in single molecule optical tweezer experiments, although the rates for RecBD1080ACD unwinding are intermediate between RecBCD and RecBΔNucCD. Surprisingly, significant pauses in DNA unwinding occur even in the absence of chi (crossover hotspot instigator) sites. We hypothesize that the nuclease domain influences the rate of DNA base pair melting, possibly allosterically and that RecBΔNucCD may mimic a post-chi state of RecBCD.
Keywords: DNA motor regulation; allostery; recombination; single molecule optical trap; transient kinetics.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
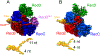

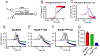

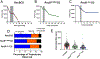
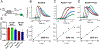

Update of
-
E. coli RecBCD Nuclease Domain Regulates Helicase Activity but not Single Stranded DNA Translocase Activity.bioRxiv [Preprint]. 2023 Oct 17:2023.10.13.561901. doi: 10.1101/2023.10.13.561901. bioRxiv. 2023. Update in: J Mol Biol. 2024 Jan 15;436(2):168381. doi: 10.1016/j.jmb.2023.168381 PMID: 37905078 Free PMC article. Updated. Preprint.
Similar articles
-
E. coli RecBCD Nuclease Domain Regulates Helicase Activity but not Single Stranded DNA Translocase Activity.bioRxiv [Preprint]. 2023 Oct 17:2023.10.13.561901. doi: 10.1101/2023.10.13.561901. bioRxiv. 2023. Update in: J Mol Biol. 2024 Jan 15;436(2):168381. doi: 10.1016/j.jmb.2023.168381 PMID: 37905078 Free PMC article. Updated. Preprint.
-
Processive DNA Unwinding by RecBCD Helicase in the Absence of Canonical Motor Translocation.J Mol Biol. 2016 Jul 31;428(15):2997-3012. doi: 10.1016/j.jmb.2016.07.002. Epub 2016 Jul 14. J Mol Biol. 2016. PMID: 27422010 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Double J Placement Methods Comparative Analysis.2023 May 30. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 May 30. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 29494060 Free Books & Documents.
-
RecBCD enzyme: mechanistic insights from mutants of a complex helicase-nuclease.Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0004123. doi: 10.1128/mmbr.00041-23. Epub 2023 Dec 4. Microbiol Mol Biol Rev. 2023. PMID: 38047637 Free PMC article. Review.
Cited by
-
Communication between DNA and nucleotide binding sites facilitates stepping by the RecBCD helicase.Nucleic Acids Res. 2024 Apr 24;52(7):3911-3923. doi: 10.1093/nar/gkae108. Nucleic Acids Res. 2024. PMID: 38364872 Free PMC article.
References
-
- Boehmer PE, Emmerson PT. The RecB subunit of the Escherichia coli RecBCD enzyme couples ATP hydrolysis to DNA unwinding. J Biol Chem. 1992;267:4981–7. - PubMed
-
- Chen HW, Ruan B, Yu M, Wang J, Julin DA. The RecD subunit of the RecBCD enzyme from Escherichia coli is a single-stranded DNA-dependent ATPase. J Biol Chem. 1997;272:10072–9. - PubMed
-
- Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

