This is a preprint.
Differential CpG methylation at Nnat in the early establishment of beta cell heterogeneity
- PMID: 38076935
- PMCID: PMC10705251
- DOI: 10.1101/2023.02.04.527050
Differential CpG methylation at Nnat in the early establishment of beta cell heterogeneity
Update in
-
Differential CpG methylation at Nnat in the early establishment of beta cell heterogeneity.Diabetologia. 2024 Jun;67(6):1079-1094. doi: 10.1007/s00125-024-06123-6. Epub 2024 Mar 21. Diabetologia. 2024. PMID: 38512414 Free PMC article.
Abstract
Aims/hypothesis: Beta cells within the pancreatic islet represent a heterogenous population wherein individual sub-groups of cells make distinct contributions to the overall control of insulin secretion. These include a subpopulation of highly-connected 'hub' cells, important for the propagation of intercellular Ca2+ waves. Functional subpopulations have also been demonstrated in human beta cells, with an altered subtype distribution apparent in type 2 diabetes. At present, the molecular mechanisms through which beta cell hierarchy is established are poorly understood. Changes at the level of the epigenome provide one such possibility which we explore here by focussing on the imprinted gene neuronatin (Nnat), which is required for normal insulin synthesis and secretion.
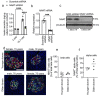
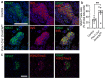
Methods: Single cell RNA-seq datasets were examined using Seurat 4.0 and ClusterProfiler running under R. Transgenic mice expressing eGFP under the control of the Nnat enhancer/promoter regions were generated for fluorescence-activated cell (FAC) sorting of beta cells and downstream analysis of CpG methylation by bisulphite and RNA sequencing, respectively. Animals deleted for the de novo methyltransferase, DNMT3A from the pancreatic progenitor stage were used to explore control of promoter methylation. Proteomics was performed using affinity purification mass spectrometry and Ca2+ dynamics explored by rapid confocal imaging of Cal-520 and Cal-590. Insulin secretion was measured using Homogeneous Time Resolved Fluorescence Imaging.
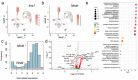
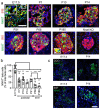
Results: Nnat mRNA was differentially expressed in a discrete beta cell population in a developmental stage- and DNA methylation (DNMT3A)-dependent manner. Thus, pseudo-time analysis of embryonic data sets demonstrated the early establishment of Nnat-positive and negative subpopulations during embryogenesis. NNAT expression is also restricted to a subset of beta cells across the human islet that is maintained throughout adult life. NNAT+ beta cells also displayed a discrete transcriptome at adult stages, representing a sub-population specialised for insulin production, reminiscent of recently-described "βHI" cells and were diminished in db/db mice. 'Hub' cells were less abundant in the NNAT+ population, consistent with epigenetic control of this functional specialization.
Conclusions/interpretation: These findings demonstrate that differential DNA methylation at Nnat represents a novel means through which beta cell heterogeneity is established during development. We therefore hypothesise that changes in methylation at this locus may thus contribute to a loss of beta cell hierarchy and connectivity, potentially contributing to defective insulin secretion in some forms of diabetes.
Keywords: Beta cell development; Ca2+; CpG methylation; connectivity; heterogeneity; identity; imprinted genes; insulin; islet; neuronatin; type 2 diabetes.
Figures




Similar articles
-
Differential CpG methylation at Nnat in the early establishment of beta cell heterogeneity.Diabetologia. 2024 Jun;67(6):1079-1094. doi: 10.1007/s00125-024-06123-6. Epub 2024 Mar 21. Diabetologia. 2024. PMID: 38512414 Free PMC article.
-
DNA methylation-mediated silencing of neuronatin (NNAT) in pig parthenogenetic fetuses.Gene. 2014 Nov 15;552(1):204-8. doi: 10.1016/j.gene.2014.09.035. Epub 2014 Sep 19. Gene. 2014. PMID: 25240791
-
Conservation of imprinting of Neuronatin (Nnat) in rabbits.Springerplus. 2015 Jun 13;4:257. doi: 10.1186/s40064-015-1054-z. eCollection 2015. Springerplus. 2015. PMID: 26090304 Free PMC article.
-
Importance of Both Imprinted Genes and Functional Heterogeneity in Pancreatic Beta Cells: Is There a Link?Int J Mol Sci. 2021 Jan 20;22(3):1000. doi: 10.3390/ijms22031000. Int J Mol Sci. 2021. PMID: 33498234 Free PMC article. Review.
-
Neuronatin gene: Imprinted and misfolded: Studies in Lafora disease, diabetes and cancer may implicate NNAT-aggregates as a common downstream participant in neuronal loss.Genomics. 2014 Feb-Mar;103(2-3):183-8. doi: 10.1016/j.ygeno.2013.12.001. Epub 2013 Dec 15. Genomics. 2014. PMID: 24345642 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
