SARS-CoV-2 infection triggers pro-atherogenic inflammatory responses in human coronary vessels
- PMID: 38076343
- PMCID: PMC10702930
- DOI: 10.1038/s44161-023-00336-5
SARS-CoV-2 infection triggers pro-atherogenic inflammatory responses in human coronary vessels
Abstract
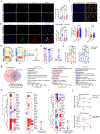
Patients with coronavirus disease 2019 (COVID-19) present increased risk for ischemic cardiovascular complications up to 1 year after infection. Although the systemic inflammatory response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection likely contributes to this increased cardiovascular risk, whether SARS-CoV-2 directly infects the coronary vasculature and attendant atherosclerotic plaques remains unknown. Here we report that SARS-CoV-2 viral RNA is detectable and replicates in coronary lesions taken at autopsy from severe COVID-19 cases. SARS-CoV-2 targeted plaque macrophages and exhibited a stronger tropism for arterial lesions than adjacent perivascular fat, correlating with macrophage infiltration levels. SARS-CoV-2 entry was increased in cholesterol-loaded primary macrophages and dependent, in part, on neuropilin-1. SARS-CoV-2 induced a robust inflammatory response in cultured macrophages and human atherosclerotic vascular explants with secretion of cytokines known to trigger cardiovascular events. Our data establish that SARS-CoV-2 infects coronary vessels, inducing plaque inflammation that could trigger acute cardiovascular complications and increase the long-term cardiovascular risk.
Conflict of interest statement
The M.S. laboratory has received unrelated research funding in sponsored research agreements from ArgenX N.V., Moderna and Phio Pharmaceuticals which has no competing interest with this work. The authors declare no other competing interests.
Figures



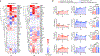

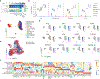

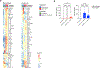





Update of
-
SARS-CoV-2 infection triggers pro-atherogenic inflammatory responses in human coronary vessels.bioRxiv [Preprint]. 2023 Aug 15:2023.08.14.553245. doi: 10.1101/2023.08.14.553245. bioRxiv. 2023. Update in: Nat Cardiovasc Res. 2023 Oct;2(10):899-916. doi: 10.1038/s44161-023-00336-5 PMID: 37645908 Free PMC article. Updated. Preprint.
Comment in
-
SARS-CoV-2 infects macrophages in coronary atherosclerotic plaques.Nat Rev Cardiol. 2023 Dec;20(12):797. doi: 10.1038/s41569-023-00949-0. Nat Rev Cardiol. 2023. PMID: 37853160 No abstract available.
Similar articles
-
SARS-CoV-2 infection triggers pro-atherogenic inflammatory responses in human coronary vessels.bioRxiv [Preprint]. 2023 Aug 15:2023.08.14.553245. doi: 10.1101/2023.08.14.553245. bioRxiv. 2023. Update in: Nat Cardiovasc Res. 2023 Oct;2(10):899-916. doi: 10.1038/s44161-023-00336-5 PMID: 37645908 Free PMC article. Updated. Preprint.
-
Spatial transcriptomic profiling of coronary endothelial cells in SARS-CoV-2 myocarditis.bioRxiv [Preprint]. 2022 Sep 26:2022.09.25.509426. doi: 10.1101/2022.09.25.509426. bioRxiv. 2022. Update in: Front Med (Lausanne). 2023 Mar 09;10:1118024. doi: 10.3389/fmed.2023.1118024 PMID: 36203548 Free PMC article. Updated. Preprint.
-
Coronavirus-19 (SARS-CoV-2) induces acute severe lung inflammation via IL-1 causing cytokine storm in COVID-19: a promising inhibitory strategy.J Biol Regul Homeost Agents. 2020 Nov-Dec;34(6):1971-1975. doi: 10.23812/20-1-E. J Biol Regul Homeost Agents. 2020. PMID: 33016027
-
Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: Lessons from SARS and MERS, and potential therapeutic interventions.Life Sci. 2020 Sep 15;257:118102. doi: 10.1016/j.lfs.2020.118102. Epub 2020 Jul 18. Life Sci. 2020. PMID: 32687918 Free PMC article. Review.
-
Cardiovascular Tropism and Sequelae of SARS-CoV-2 Infection.Viruses. 2022 May 25;14(6):1137. doi: 10.3390/v14061137. Viruses. 2022. PMID: 35746609 Free PMC article. Review.
Cited by
-
Assessing the Potential of an Enzymatically Liberated Salmon Oil to Support Immune Health Recovery from Acute SARS-CoV-2 Infection via Change in the Expression of Cytokine, Chemokine and Interferon-Related Genes.Int J Mol Sci. 2024 Jun 24;25(13):6917. doi: 10.3390/ijms25136917. Int J Mol Sci. 2024. PMID: 39000027 Free PMC article. Clinical Trial.
-
Innate Immune Dysregulations and Cross Talk in COVID-19: Novel Players in Atherogenesis.Arterioscler Thromb Vasc Biol. 2024 Oct;44(10):2223-2225. doi: 10.1161/ATVBAHA.124.321415. Epub 2024 Aug 8. Arterioscler Thromb Vasc Biol. 2024. PMID: 39114915 No abstract available.
-
Reduced HDL-cholesterol in long COVID-19: A key metabolic risk factor tied to disease severity.Clinics (Sao Paulo). 2024 Mar 28;79:100344. doi: 10.1016/j.clinsp.2024.100344. eCollection 2024. Clinics (Sao Paulo). 2024. PMID: 38552385 Free PMC article.
-
Emerging Roles of Exosomes in Stroke Therapy.Int J Mol Sci. 2024 Jun 13;25(12):6507. doi: 10.3390/ijms25126507. Int J Mol Sci. 2024. PMID: 38928214 Free PMC article. Review.
-
Enhancing Wound Healing and Anti-Inflammatory Effects by Combination of CIGB-258 and Apolipoprotein A-I against Carboxymethyllysine Toxicity in Zebrafish: Insights into Structural Stabilization and Antioxidant Properties.Antioxidants (Basel). 2024 Aug 28;13(9):1049. doi: 10.3390/antiox13091049. Antioxidants (Basel). 2024. PMID: 39334708 Free PMC article.
References
-
- Katsoularis I, Fonseca-Rodriguez O, Farrington P, Lindmark K & Fors Connolly AM Risk of acute myocardial infarction and ischaemic stroke following COVID-19 in Sweden: a self-controlled case series and matched cohort study. Lancet 398, 599–607 (2021). 10.1016/S0140-6736(21)00896-5 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
