COVID-19 ORF3a Viroporin-Influenced Common and Unique Cellular Signaling Cascades in Lung, Heart, and the Brain Choroid Plexus Organoids with Additional Enriched MicroRNA Network Analyses for Lung and the Brain Tissues
- PMID: 38075756
- PMCID: PMC10701872
- DOI: 10.1021/acsomega.3c06485
COVID-19 ORF3a Viroporin-Influenced Common and Unique Cellular Signaling Cascades in Lung, Heart, and the Brain Choroid Plexus Organoids with Additional Enriched MicroRNA Network Analyses for Lung and the Brain Tissues
Abstract
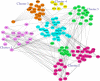
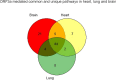
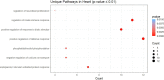
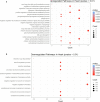
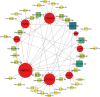
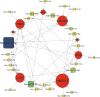
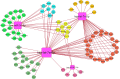
Tissue-specific implications of SARS-CoV-2-encoded accessory proteins are not fully understood. SARS-CoV-2 infection can severely affect three major organs-the heart, lungs, and brain. We analyzed SARS-CoV-2 ORF3a interacting host proteins in these three major organs. Furthermore, we identified common and unique interacting host proteins and their targeting miRNAs (lung and brain) and delineated associated biological processes by reanalyzing RNA-seq data from the brain (COVID-19-infected/uninfected choroid plexus organoid study), lung tissue from COVID-19 patients/healthy subjects, and cardiomyocyte cells-based transcriptomics analyses. Our in silico studies showed ORF3a interacting proteins could vary depending upon tissues. The number of unique ORF3a interacting proteins in the brain, lungs, and heart were 10, 7, and 1, respectively. Though common pathways influenced by SARS-CoV-2 infection were more, unique 21 brain and 7 heart pathways were found. One unique pathway for the heart was negative regulation of calcium ion transport. Reported observations of COVID-19 patients with a history of hypertension taking calcium channel blockers (CCBs) or dihydropyridine CCBs had an elevated rate of intubation or increased rate of intubation/death, respectively. Also, the likelihood of hospitalization of chronic CCB users with COVID-19 was greater in comparison to long-term angiotensin-converting enzyme inhibitors/angiotensin receptor blockers users. Further studies are necessary to confirm this. miRNA analysis of ORF3a interacting proteins in the brain and lungs revealed 3 of 37 brain miRNAs and 1 of 25 lung miRNAs with high degree and betweenness indicating their significance as hubs in the interaction network. Our study could help in identifying potential tissue-specific COVID-19 drug/drug repurposing targets.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





Similar articles
-
Association of renin-angiotensin-aldosterone system inhibition with Covid-19 hospitalization and all-cause mortality in the UK biobank.Br J Clin Pharmacol. 2022 Jun;88(6):2830-2842. doi: 10.1111/bcp.15192. Epub 2022 Jan 27. Br J Clin Pharmacol. 2022. PMID: 34935181
-
Antihypertensive Drugs and COVID-19 Risk: A Cohort Study of 2 Million Hypertensive Patients.Hypertension. 2021 Mar 3;77(3):833-842. doi: 10.1161/HYPERTENSIONAHA.120.16314. Epub 2021 Jan 11. Hypertension. 2021. PMID: 33423528 Free PMC article.
-
The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.bioRxiv [Preprint]. 2022 Sep 3:2022.09.02.506428. doi: 10.1101/2022.09.02.506428. bioRxiv. 2022. Update in: Elife. 2023 Jan 25;12:e84477. doi: 10.7554/eLife.84477. PMID: 36263072 Free PMC article. Updated. Preprint.
-
SARS-CoV-2 pandemic and research gaps: Understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy.Theranostics. 2020 Jun 12;10(16):7448-7464. doi: 10.7150/thno.48076. eCollection 2020. Theranostics. 2020. PMID: 32642005 Free PMC article. Review.
-
Human brain organoids to explore SARS-CoV-2-induced effects on the central nervous system.Rev Med Virol. 2023 Mar;33(2):e2430. doi: 10.1002/rmv.2430. Epub 2023 Feb 15. Rev Med Virol. 2023. PMID: 36790825 Review.
References
-
- Larsen H. D.; Fonager J.; Lomholt F. K.; Dalby T.; Benedetti G.; Kristensen B.; Urth T. R.; Rasmussen M.; Lassaunière R.; Rasmussen T. B.; Strandbygaard B.; Lohse L.; Chaine M.; Møller K. L.; Berthelsen A.-S. N.; Nørgaard S. K.; Sönksen U. W.; Boklund A. E.; Hammer A. S.; Belsham G. J.; Krause T. G.; Mortensen S.; Bøtner A.; Fomsgaard A.; Mølbak K.. Preliminary Report of an Outbreak of SARS-CoV-2 in Mink and Mink Farmers Associated with Community Spread, Denmark, June to November 2020. Eurosurveillance 2021, 26( (5), ), pii=2100009.10.2807/1560-7917.ES.2021.26.5.210009 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

