Exploiting autophagy balance in T and NK cells as a new strategy to implement adoptive cell therapies
- PMID: 38071322
- PMCID: PMC10709869
- DOI: 10.1186/s12943-023-01893-w
Exploiting autophagy balance in T and NK cells as a new strategy to implement adoptive cell therapies
Abstract
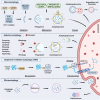
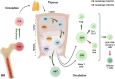
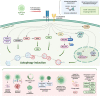
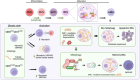
Autophagy is an essential cellular homeostasis pathway initiated by multiple stimuli ranging from nutrient deprivation to viral infection, playing a key role in human health and disease. At present, a growing number of evidence suggests a role of autophagy as a primitive innate immune form of defense for eukaryotic cells, interacting with components of innate immune signaling pathways and regulating thymic selection, antigen presentation, cytokine production and T/NK cell homeostasis. In cancer, autophagy is intimately involved in the immunological control of tumor progression and response to therapy. However, very little is known about the role and impact of autophagy in T and NK cells, the main players in the active fight against infections and tumors. Important questions are emerging: what role does autophagy play on T/NK cells? Could its modulation lead to any advantages? Could specific targeting of autophagy on tumor cells (blocking) and T/NK cells (activation) be a new intervention strategy? In this review, we debate preclinical studies that have identified autophagy as a key regulator of immune responses by modulating the functions of different immune cells and discuss the redundancy or diversity among the subpopulations of both T and NK cells in physiologic context and in cancer.
Keywords: Autophagy; Effector cells; Metabolism; Mitophagy; T and NK development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Exploiting Human NK Cells in Tumor Therapy.Front Immunol. 2020 Jan 17;10:3013. doi: 10.3389/fimmu.2019.03013. eCollection 2019. Front Immunol. 2020. PMID: 32010130 Free PMC article. Review.
-
CD8 T cells in innate immune responses: using STAT4-dependent but antigen-independent pathways to gamma interferon during viral infection.mBio. 2014 Oct 21;5(5):e01978-14. doi: 10.1128/mBio.01978-14. mBio. 2014. PMID: 25336459 Free PMC article.
-
The dual-functional capability of cytokine-induced killer cells and application in tumor immunology.Hum Immunol. 2015 May;76(5):385-91. doi: 10.1016/j.humimm.2014.09.021. Epub 2014 Oct 8. Hum Immunol. 2015. PMID: 25305457 Review.
-
Involvement of autophagy in NK cell development and function.Autophagy. 2017 Mar 4;13(3):633-636. doi: 10.1080/15548627.2016.1274486. Epub 2017 Jan 19. Autophagy. 2017. PMID: 28103115 Free PMC article.
-
Positive & Negative Roles of Innate Effector Cells in Controlling Cancer Progression.Front Immunol. 2018 Sep 21;9:1990. doi: 10.3389/fimmu.2018.01990. eCollection 2018. Front Immunol. 2018. PMID: 30298063 Free PMC article. Review.
Cited by
-
Transition to a mesenchymal state in neuroblastoma may be characterized by a high expression of GD2 and by the acquisition of immune escape from NK cells.Front Immunol. 2024 Apr 26;15:1382931. doi: 10.3389/fimmu.2024.1382931. eCollection 2024. Front Immunol. 2024. PMID: 38736882 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

