PML Body Biogenesis: A Delicate Balance of Interactions
- PMID: 38069029
- PMCID: PMC10705990
- DOI: 10.3390/ijms242316702
PML Body Biogenesis: A Delicate Balance of Interactions
Abstract
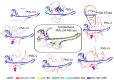
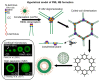
PML bodies are subnuclear protein complexes that play a crucial role in various physiological and pathological cellular processes. One of the general structural proteins of PML bodies is a member of the tripartite motif (TRIM) family-promyelocytic leukemia protein (PML). It is known that PML interacts with over a hundred partners, and the protein itself is represented by several major isoforms, differing in their variable and disordered C-terminal end due to alternative splicing. Despite nearly 30 years of research, the mechanisms underlying PML body formation and the role of PML proteins in this process remain largely unclear. In this review, we examine the literature and highlight recent progress in this field, with a particular focus on understanding the role of individual domains of the PML protein, its post-translational modifications, and polyvalent nonspecific interactions in the formation of PML bodies. Additionally, based on the available literature, we propose a new hypothetical model of PML body formation.
Keywords: PML-bodies; SUMO/SIM; TRIM domain; biomolecular condensates; intrinsically disordered protein; liquid–liquid phase separation; membraneless organelle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
On the Prevalence and Roles of Proteins Undergoing Liquid-Liquid Phase Separation in the Biogenesis of PML-Bodies.Biomolecules. 2023 Dec 18;13(12):1805. doi: 10.3390/biom13121805. Biomolecules. 2023. PMID: 38136675 Free PMC article.
-
PML Nuclear bodies: the cancer connection and beyond.Nucleus. 2024 Dec;15(1):2321265. doi: 10.1080/19491034.2024.2321265. Epub 2024 Feb 27. Nucleus. 2024. PMID: 38411156 Free PMC article. Review.
-
New Evidence of the Importance of Weak Interactions in the Formation of PML-Bodies.Int J Mol Sci. 2022 Jan 30;23(3):1613. doi: 10.3390/ijms23031613. Int J Mol Sci. 2022. PMID: 35163537 Free PMC article.
-
Contribution of the C-terminal regions of promyelocytic leukemia protein (PML) isoforms II and V to PML nuclear body formation.J Biol Chem. 2012 Aug 31;287(36):30729-42. doi: 10.1074/jbc.M112.374769. Epub 2012 Jul 7. J Biol Chem. 2012. PMID: 22773875 Free PMC article.
-
The promyelocytic leukemia nuclear body: sites of activity?Biochem Cell Biol. 2002;80(3):301-10. doi: 10.1139/o02-079. Biochem Cell Biol. 2002. PMID: 12123283 Review.
Cited by
-
Multifunctional histone variants in genome function.Nat Rev Genet. 2025 Feb;26(2):82-104. doi: 10.1038/s41576-024-00759-1. Epub 2024 Aug 13. Nat Rev Genet. 2025. PMID: 39138293 Review.
-
Characterization and functional analysis of chicken promyelocytic leukemia protein.Poult Sci. 2024 Dec;103(12):104272. doi: 10.1016/j.psj.2024.104272. Epub 2024 Aug 28. Poult Sci. 2024. PMID: 39293264 Free PMC article.
References
-
- Kakizuka A., Miller W.H., Umesono K., Warrell R.P., Frankel S.R., Murty V.V.V.S., Dmitrovsky E., Evans R.M. Chromosomal Translocation t(15;17) in Human Acute Promyelocytic Leukemia Fuses RARα with a Novel Putative Transcription Factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-C. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

