A Natural Compound Containing a Disaccharide Structure of Glucose and Rhamnose Identified as Potential N-Glycanase 1 (NGLY1) Inhibitors
- PMID: 38067490
- PMCID: PMC10707914
- DOI: 10.3390/molecules28237758
A Natural Compound Containing a Disaccharide Structure of Glucose and Rhamnose Identified as Potential N-Glycanase 1 (NGLY1) Inhibitors
Abstract
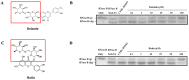
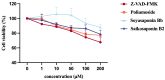
N-glycanase 1 (NGLY1) is an essential enzyme involved in the deglycosylation of misfolded glycoproteins through the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway, which could hydrolyze N-glycan from N-glycoprotein or N-glycopeptide in the cytosol. Recent studies indicated that NGLY1 inhibition is a potential novel drug target for antiviral therapy. In this study, structure-based virtual analysis was applied to screen candidate NGLY1 inhibitors from 2960 natural compounds. Three natural compounds, Poliumoside, Soyasaponin Bb, and Saikosaponin B2 showed significantly inhibitory activity of NGLY1, isolated from traditional heat-clearing and detoxifying Chinese herbs. Furthermore, the core structural motif of the three NGLY1 inhibitors was a disaccharide structure with glucose and rhamnose, which might exert its action by binding to important active sites of NGLY1, such as Lys238 and Trp244. In traditional Chinese medicine, many compounds containing this disaccharide structure probably targeted NGLY1. This study unveiled the leading compound of NGLY1 inhibitors with its core structure, which could guide future drug development.
Keywords: N-glycanase 1 (NGLY1); NGLY1 inhibitor; natural compound; structure-based virtual screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
The cytoplasmic peptide:N-glycanase (NGLY1) - Structure, expression and cellular functions.Gene. 2016 Feb 10;577(1):1-7. doi: 10.1016/j.gene.2015.11.021. Epub 2015 Nov 30. Gene. 2016. PMID: 26611529 Free PMC article. Review.
-
Repurposing of Proton Pump Inhibitors as first identified small molecule inhibitors of endo-β-N-acetylglucosaminidase (ENGase) for the treatment of NGLY1 deficiency, a rare genetic disease.Bioorg Med Chem Lett. 2017 Jul 1;27(13):2962-2966. doi: 10.1016/j.bmcl.2017.05.010. Epub 2017 May 5. Bioorg Med Chem Lett. 2017. PMID: 28512024 Free PMC article.
-
Comprehensive Analysis of the Structure and Function of Peptide:N-Glycanase 1 and Relationship with Congenital Disorder of Deglycosylation.Nutrients. 2022 Apr 19;14(9):1690. doi: 10.3390/nu14091690. Nutrients. 2022. PMID: 35565658 Free PMC article. Review.
-
Physiological importance of NGLY1, as revealed by rodent model analyses.J Biochem. 2022 Feb 21;171(2):161-167. doi: 10.1093/jb/mvab101. J Biochem. 2022. PMID: 34580715 Review.
-
Endo-β-N-acetylglucosaminidase forms N-GlcNAc protein aggregates during ER-associated degradation in Ngly1-defective cells.Proc Natl Acad Sci U S A. 2015 Feb 3;112(5):1398-403. doi: 10.1073/pnas.1414593112. Epub 2015 Jan 20. Proc Natl Acad Sci U S A. 2015. PMID: 25605922 Free PMC article.
References
-
- Tomlin F.M., Gerling-Driessen U.I.M., Liu Y.C., Flynn R.A., Vangala J.R., Lentz C.S., Clauder-Muenster S., Jakob P., Mueller W.F., Ordoñez-Rueda D., et al. Inhibition of NGLY1 Inactivates the Transcription Factor Nrf1 and Potentiates Proteasome Inhibitor Cytotoxicity. ACS Cent. Sci. 2017;3:1143–1155. doi: 10.1021/acscentsci.7b00224. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

