Epigenetic Regulation in Oral Squamous Cell Carcinoma Microenvironment: A Comprehensive Review
- PMID: 38067304
- PMCID: PMC10705512
- DOI: 10.3390/cancers15235600
Epigenetic Regulation in Oral Squamous Cell Carcinoma Microenvironment: A Comprehensive Review
Abstract
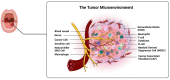
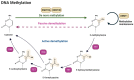
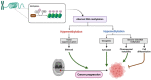
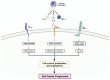
Oral squamous cell carcinoma (OSCC) is a prevalent and significant type of oral cancer that has far-reaching health implications worldwide. Epigenetics, a field focused on studying heritable changes in gene expression without modifying DNA sequence, plays a pivotal role in OSCC. Epigenetic changes, encompassing DNA methylation, histone modifications, and miRNAs, exert control over gene activity and cellular characteristics. In OSCC, aberrant DNA methylation of tumor suppressor genes (TSG) leads to their inactivation, subsequently facilitating tumor growth. As a result, distinct patterns of gene methylation hold promise as valuable biomarkers for the detection of OSCC. Oral cancer treatment typically involves surgery, radiation therapy, and chemotherapy, but even with these treatments, cancer cells cannot be effectively targeted and destroyed. Researchers are therefore exploring new methods to target and eliminate cancer cells. One promising approach is the use of epigenetic modifiers, such as DNA methyltransferase (DNMT) inhibitors and histone deacetylase (HDAC) inhibitors, which have been shown to modify abnormal epigenetic patterns in OSCC cells, leading to the reactivation of TSGs and the suppression of oncogenes. As a result, epigenetic-targeted therapies have the potential to directly alter gene expression and minimize side effects. Several studies have explored the efficacy of such therapies in the treatment of OSCC. Although studies have investigated the efficacy of epigenetic therapies, challenges in identifying reliable biomarkers and developing effective combination treatments are acknowledged. Of note, epigenetic mechanisms play a significant role in drug resistance in OSCC and other cancers. Aberrant DNA methylation can silence tumor suppressor genes, while alterations in histone modifications and chromatin remodeling affect gene expression related to drug metabolism and cell survival. Thus, understanding and targeting these epigenetic processes offer potential strategies to overcome drug resistance and improve the efficacy of cancer treatments in OSCC. This comprehensive review focuses on the complex interplay between epigenetic alterations and OSCC cells. This will involve a deep dive into the mechanisms underlying epigenetic modifications and their impact on OSCC, including its initiation, progression, and metastasis. Furthermore, this review will present the role of epigenetics in the treatment and diagnosis of OSCC.
Keywords: DNA methylation; drug resistance; epigenetic; oral squamous cell carcinoma; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Promoter Hypermethylation of LATS2 Gene in Oral Squamous Cell Carcinoma (OSCC) Among North Indian Population.Asian Pac J Cancer Prev. 2020 May 1;21(5):1283-1287. doi: 10.31557/APJCP.2020.21.5.1283. Asian Pac J Cancer Prev. 2020. PMID: 32458634 Free PMC article.
-
Aberrantly hypermethylated tumor suppressor genes were identified in oral squamous cell carcinoma (OSCC).Clin Epigenetics. 2019 Aug 12;11(1):116. doi: 10.1186/s13148-019-0715-0. Clin Epigenetics. 2019. PMID: 31405379 Free PMC article.
-
Impact of Epigenetic Alterations in the Development of Oral Diseases.Curr Med Chem. 2021;28(6):1091-1103. doi: 10.2174/0929867327666200114114802. Curr Med Chem. 2021. PMID: 31942842 Review.
-
Epigenetics in oral squamous cell carcinoma.J Oral Maxillofac Pathol. 2017 May-Aug;21(2):252-259. doi: 10.4103/jomfp.JOMFP_150_17. J Oral Maxillofac Pathol. 2017. PMID: 28932035 Free PMC article. Review.
-
Histone lysine methyltransferase SMYD3 promotes oral squamous cell carcinoma tumorigenesis via H3K4me3-mediated HMGA2 transcription.Clin Epigenetics. 2023 May 26;15(1):92. doi: 10.1186/s13148-023-01506-9. Clin Epigenetics. 2023. PMID: 37237385 Free PMC article.
Cited by
-
Assessing DNA methylation of ATG 5 and MAP1LC3Av1 gene in oral squamous cell carcinoma and oral leukoplakia- a cross sectional study.J Oral Biol Craniofac Res. 2024 Sep-Oct;14(5):534-539. doi: 10.1016/j.jobcr.2024.07.001. Epub 2024 Jul 6. J Oral Biol Craniofac Res. 2024. PMID: 39070885 Free PMC article.
-
Identification of Stage-Specific microRNAs that Govern the Early Stages of Sequential Oral Oncogenesis by Strategically Bridging Human Genetics with Epigenetics and Utilizing an Animal Model.Int J Mol Sci. 2024 Jul 12;25(14):7642. doi: 10.3390/ijms25147642. Int J Mol Sci. 2024. PMID: 39062890 Free PMC article.
References
-
- Nicolette Salmon H., Jan-MichaÉL H., Jamshid J., Miranda M.J., Lars S. Viral and Molecular Aspects of Oral Cancer. Anticancer Res. 2012;32:4201. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

