Inhibition of human cytomegalovirus replication by interferon alpha can involve multiple anti-viral factors
- PMID: 38063292
- PMCID: PMC10770924
- DOI: 10.1099/jgv.0.001929
Inhibition of human cytomegalovirus replication by interferon alpha can involve multiple anti-viral factors
Abstract
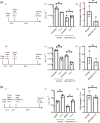
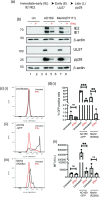
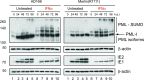
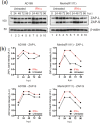
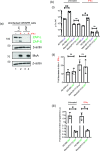
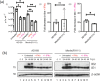
The shortcomings of current direct-acting anti-viral therapy against human cytomegalovirus (HCMV) has led to interest in host-directed therapy. Here we re-examine the use of interferon proteins to inhibit HCMV replication utilizing both high and low passage strains of HCMV. Pre-treatment of cells with interferon alpha (IFNα) was required for robust and prolonged inhibition of both low and high passage HCMV strains, with no obvious toxicity, and was associated with an increased anti-viral state in HCMV-infected cells. Pre-treatment of cells with IFNα led to poor expression of HCMV immediate-early proteins from both high and low passage strains, which was associated with the presence of the anti-viral factor SUMO-PML. Inhibition of HCMV replication in the presence of IFNα involving ZAP proteins was HCMV strain-dependent, wherein a high passage HCMV strain was obviously restricted by ZAP and a low passage strain was not. This suggested that strain-specific combinations of anti-viral factors were involved in inhibition of HCMV replication in the presence of IFNα. Overall, this work further supports the development of strategies involving IFNα that may be useful to inhibit HCMV replication and highlights the complexity of the anti-viral response to HCMV in the presence of IFNα.
Keywords: human cytomegalovirus interferon alpha PML ZAP.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
A broadly neutralizing human monoclonal antibody generated from transgenic mice immunized with HCMV particles limits virus infection and proliferation.J Virol. 2024 Jul 23;98(7):e0021324. doi: 10.1128/jvi.00213-24. Epub 2024 Jun 4. J Virol. 2024. PMID: 38832789 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Antioxidants for female subfertility.Cochrane Database Syst Rev. 2020 Aug 27;8(8):CD007807. doi: 10.1002/14651858.CD007807.pub4. Cochrane Database Syst Rev. 2020. PMID: 32851663 Free PMC article.
-
Antioxidants for female subfertility.Cochrane Database Syst Rev. 2017 Jul 28;7(7):CD007807. doi: 10.1002/14651858.CD007807.pub3. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Aug 27;8:CD007807. doi: 10.1002/14651858.CD007807.pub4 PMID: 28752910 Free PMC article. Updated. Review.
Cited by
-
cGAS-STING-TBK1 Signaling Promotes Valproic Acid-Responsive Human Cytomegalovirus Immediate-Early Transcription during Infection of Incompletely Differentiated Myeloid Cells.Viruses. 2024 May 30;16(6):877. doi: 10.3390/v16060877. Viruses. 2024. PMID: 38932169 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

