Spatial regulation of DNA damage tolerance protein Rad5 interconnects genome stability maintenance and proteostasis networks
- PMID: 38055836
- PMCID: PMC10853803
- DOI: 10.1093/nar/gkad1176
Spatial regulation of DNA damage tolerance protein Rad5 interconnects genome stability maintenance and proteostasis networks
Abstract
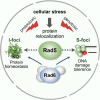
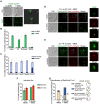
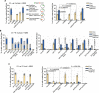
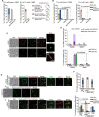
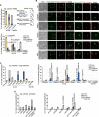
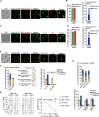
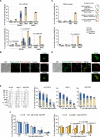
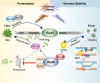
The Rad5/HLTF protein has a central role in the tolerance to DNA damage by mediating an error-free mode of bypassing unrepaired DNA lesions, and is therefore critical for the maintenance of genome stability. We show in this work that, following cellular stress, Rad5 is regulated by relocalization into two types of nuclear foci that coexist within the same cell, which we termed 'S' and 'I'. Rad5 S-foci form in response to genotoxic stress and are associated with Rad5's function in maintaining genome stability, whereas I-foci form in the presence of proteotoxic stress and are related to Rad5's own proteostasis. Rad5 accumulates into S-foci at DNA damage tolerance sites by liquid-liquid phase separation, while I-foci constitute sites of chaperone-mediated sequestration of Rad5 at the intranuclear quality control compartment (INQ). Relocalization of Rad5 into each type of foci involves different pathways and recruitment mechanisms, but in both cases is driven by the evolutionarily conserved E2 ubiquitin-conjugating enzyme Rad6. This coordinated differential relocalization of Rad5 interconnects DNA damage response and proteostasis networks, highlighting the importance of studying these homeostasis mechanisms in tandem. Spatial regulation of Rad5 under cellular stress conditions thus provides a useful biological model to study cellular homeostasis as a whole.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Rad5 plays a major role in the cellular response to DNA damage during chromosome replication.Cell Rep. 2014 Oct 23;9(2):460-8. doi: 10.1016/j.celrep.2014.09.005. Epub 2014 Oct 9. Cell Rep. 2014. PMID: 25310987
-
Post-replication repair suppresses duplication-mediated genome instability.PLoS Genet. 2010 May 6;6(5):e1000933. doi: 10.1371/journal.pgen.1000933. PLoS Genet. 2010. PMID: 20463880 Free PMC article.
-
RAD6-RAD18-RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light.Nature. 2009 Jan 29;457(7229):612-5. doi: 10.1038/nature07580. Epub 2008 Dec 14. Nature. 2009. PMID: 19079240
-
Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance.DNA Repair (Amst). 2010 Mar 2;9(3):257-67. doi: 10.1016/j.dnarep.2009.12.013. Epub 2010 Jan 21. DNA Repair (Amst). 2010. PMID: 20096653 Review.
-
Mechanistic Insights into the Multiple Activities of the Rad5 Family of Enzymes.J Mol Biol. 2022 May 30;434(10):167581. doi: 10.1016/j.jmb.2022.167581. Epub 2022 Apr 7. J Mol Biol. 2022. PMID: 35398319 Review.
Cited by
-
Dynamics of DNA damage-induced nuclear inclusions are regulated by SUMOylation of Btn2.Nat Commun. 2024 Apr 13;15(1):3215. doi: 10.1038/s41467-024-47615-8. Nat Commun. 2024. PMID: 38615096 Free PMC article.
References
-
- Aguilera A., García-Muse T.. Causes of genome instability. Annu. Rev. Genet. 2013; 47:1–32. - PubMed
-
- Hipp M.S., Kasturi P., Hartl F.U.. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019; 20:421–435. - PubMed
-
- Huiting W., Bergink S.. Locked in a vicious cycle: the connection between genomic instability and a loss of protein homeostasis. Genome Instab. Dis. 2021; 2:1–23.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

