Intrinsic factors and CD1d1 but not CD1d2 expression levels control invariant natural killer T cell subset differentiation
- PMID: 38040679
- PMCID: PMC10692182
- DOI: 10.1038/s41467-023-43424-7
Intrinsic factors and CD1d1 but not CD1d2 expression levels control invariant natural killer T cell subset differentiation
Abstract
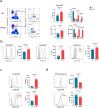
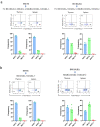
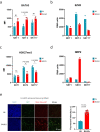
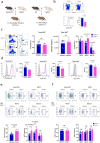
Invariant natural killer T (NKT) cell subsets are defined based on their cytokine-production profiles and transcription factors. Their distribution is different in C57BL/6 (B6) and BALB/c mice, with a bias for NKT1 and NKT2/NKT17 subsets, respectively. Here, we show that the non-classical class I-like major histocompatibility complex CD1 molecules CD1d2, expressed in BALB/c and not in B6 mice, could not account for this difference. We find however that NKT cell subset distribution is intrinsic to bone marrow derived NKT cells, regardless of syngeneic CD1d-ligand recognition, and that multiple intrinsic factors are likely involved. Finally, we find that CD1d expression levels in combination with T cell antigen receptor signal strength could also influence NKT cell distribution and function. Overall, this study indicates that CD1d-mediated TCR signals and other intrinsic signals integrate to influence strain-specific NKT cell differentiation programs and subset distributions.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Differing roles of CD1d2 and CD1d1 proteins in type I natural killer T cell development and function.Proc Natl Acad Sci U S A. 2018 Feb 6;115(6):E1204-E1213. doi: 10.1073/pnas.1716669115. Epub 2018 Jan 19. Proc Natl Acad Sci U S A. 2018. PMID: 29351991 Free PMC article.
-
Defining a novel subset of CD1d-dependent type II natural killer T cells using natural killer cell-associated markers.Scand J Immunol. 2019 Sep;90(3):e12794. doi: 10.1111/sji.12794. Epub 2019 Jun 26. Scand J Immunol. 2019. PMID: 31141185 Free PMC article.
-
Human invariant V alpha 24-J alpha Q TCR supports the development of CD1d-dependent NK1.1+ and NK1.1- T cells in transgenic mice.J Immunol. 2003 Mar 1;170(5):2390-8. doi: 10.4049/jimmunol.170.5.2390. J Immunol. 2003. PMID: 12594262
-
The Role of Adaptor Proteins in the Biology of Natural Killer T (NKT) Cells.Front Immunol. 2019 Jun 25;10:1449. doi: 10.3389/fimmu.2019.01449. eCollection 2019. Front Immunol. 2019. PMID: 31293596 Free PMC article. Review.
-
Invariant NKT and NKT-like lymphocytes: two different T cell subsets that are differentially affected by ageing.Exp Gerontol. 2007 Aug;42(8):703-8. doi: 10.1016/j.exger.2007.05.002. Epub 2007 May 21. Exp Gerontol. 2007. PMID: 17604928 Review.
Cited by
-
Innate-like T cell subset commitment in the murine thymus is independent of TCR characteristics and occurs during proliferation.Proc Natl Acad Sci U S A. 2024 Apr 2;121(14):e2311348121. doi: 10.1073/pnas.2311348121. Epub 2024 Mar 26. Proc Natl Acad Sci U S A. 2024. PMID: 38530897 Free PMC article.
-
CD1 molecules: Beyond antigen presentation.Mol Immunol. 2024 Jun;170:1-8. doi: 10.1016/j.molimm.2024.03.011. Epub 2024 Apr 4. Mol Immunol. 2024. PMID: 38579449 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

