GC heterogeneity reveals sequence-structures evolution of angiosperm ITS2
- PMID: 38036992
- PMCID: PMC10691020
- DOI: 10.1186/s12870-023-04634-9
GC heterogeneity reveals sequence-structures evolution of angiosperm ITS2
Abstract
Background: Despite GC variation constitutes a fundamental element of genome and species diversity, the precise mechanisms driving it remain unclear. The abundant sequence data available for the ITS2, a commonly employed phylogenetic marker in plants, offers an exceptional resource for exploring the GC variation across angiosperms.
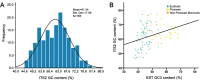
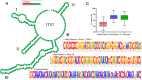
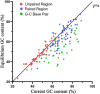
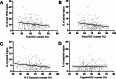
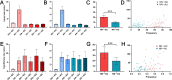
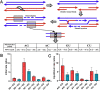
Results: A comprehensive selection of 8666 species, comprising 165 genera, 63 families, and 30 orders were used for the analyses. The alignment of ITS2 sequence-structures and partitioning of secondary structures into paired and unpaired regions were performed using 4SALE. Substitution rates and frequencies among GC base-pairs in the paired regions of ITS2 were calculated using RNA-specific models in the PHASE package. The results showed that the distribution of ITS2 GC contents on the angiosperm phylogeny was heterogeneous, but their increase was generally associated with ITS2 sequence homogenization, thereby supporting the occurrence of GC-biased gene conversion (gBGC) during the concerted evolution of ITS2. Additionally, the GC content in the paired regions of the ITS2 secondary structure was significantly higher than that of the unpaired regions, indicating the selection of GC for thermodynamic stability. Furthermore, the RNA substitution models demonstrated that base-pair transformations favored both the elevation and fixation of GC in the paired regions, providing further support for gBGC.
Conclusions: Our findings highlight the significance of secondary structure in GC investigation, which demonstrate that both gBGC and structure-based selection are influential factors driving angiosperm ITS2 GC content.
Keywords: GC-biased gene conversion; ITS2 content; Secondary structure; Thermodynamic stability.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Molecular identification and studies on genetic diversity and structure-related GC heterogeneity of Spatholobus Suberectus based on ITS2.Sci Rep. 2024 Oct 9;14(1):23523. doi: 10.1038/s41598-024-75763-w. Sci Rep. 2024. PMID: 39384849 Free PMC article.
-
Structure-Based GC Investigation Sheds New Light on ITS2 Evolution in Corydalis Species.Int J Mol Sci. 2023 Apr 23;24(9):7716. doi: 10.3390/ijms24097716. Int J Mol Sci. 2023. PMID: 37175423 Free PMC article.
-
Adenine·cytosine substitutions are an alternative pathway of compensatory mutation in angiosperm ITS2.RNA. 2020 Feb;26(2):209-217. doi: 10.1261/rna.072660.119. Epub 2019 Nov 20. RNA. 2020. PMID: 31748405 Free PMC article.
-
GC content evolution in coding regions of angiosperm genomes: a unifying hypothesis.Trends Genet. 2014 Jul;30(7):263-70. doi: 10.1016/j.tig.2014.05.002. Epub 2014 Jun 7. Trends Genet. 2014. PMID: 24916172 Review.
-
GC-biased gene conversion links the recombination landscape and demography to genomic base composition: GC-biased gene conversion drives genomic base composition across a wide range of species.Bioessays. 2015 Dec;37(12):1317-26. doi: 10.1002/bies.201500058. Epub 2015 Oct 7. Bioessays. 2015. PMID: 26445215 Review.
Cited by
-
DNA barcoding combined with high-resolution melting analysis to discriminate rhubarb species and its traditional Chinese patent medicines.Front Pharmacol. 2024 Jun 14;15:1371890. doi: 10.3389/fphar.2024.1371890. eCollection 2024. Front Pharmacol. 2024. PMID: 38948467 Free PMC article.
-
Deciphering the role of tryptophan metabolism-associated genes ECHS1 and ALDH2 in gastric cancer: implications for tumor immunity and personalized therapy.Front Immunol. 2024 Sep 12;15:1460308. doi: 10.3389/fimmu.2024.1460308. eCollection 2024. Front Immunol. 2024. PMID: 39328412 Free PMC article.
-
Molecular identification and studies on genetic diversity and structure-related GC heterogeneity of Spatholobus Suberectus based on ITS2.Sci Rep. 2024 Oct 9;14(1):23523. doi: 10.1038/s41598-024-75763-w. Sci Rep. 2024. PMID: 39384849 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

