Depletion of lncRNA MEG3 Ameliorates Imatinib-Induced Injury of Cardiomyocytes via Regulating miR-129-5p/HMGB1 Axis
- PMID: 38028435
- PMCID: PMC10673670
- DOI: 10.1155/2023/1108280
Depletion of lncRNA MEG3 Ameliorates Imatinib-Induced Injury of Cardiomyocytes via Regulating miR-129-5p/HMGB1 Axis
Abstract
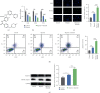
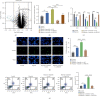
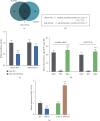
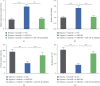
Imatinib is a classical targeted drug to treat chronic myeloid leukemia (CML). However, it shows cardiotoxicity, which limits its clinical application. Long noncoding RNA (lncRNA) maternally expressed gene 3 (MEG3) shows proapoptotic properties in human cells. This study is performed to investigate whether targeting MEG3 can attenuate imatinib-mediated cardiotoxicity to cardiomyocytes. In this work, H9c2 cells were divided into four groups: control group, hypoxia group, hypoxia + imatinib, and hypoxia + imatinib + MEG3 knockdown group. MEG3 and microRNA-129-5p (miR-129-5p) expression levels were detected by the quantitative real-time PCR (qRT-PCR). The viability and apoptosis of H9c2 cells were then evaluated by cell counting kit-8 (CCK-8), flow cytometry, and TUNEL assays. The targeting relationships between MEG3 and miR-129-5p, between miR-129-5p and high-mobility group box 1 (HMBG1), were validated by dual-luciferase reporter assay and RNA Immunoprecipitation (RIP) assay. The protein expression level of HMGB1 was detected by western blot. It was revealed that, Imatinib-inhibited cell viability and aggravated the apoptosis of H9c2 cells cultured in hypoxic condition, and MEG3 knockdown significantly counteracted this effect. MiR-129-5p was a downstream target of MEG3 and it directly targeted HMGB1, and knockdown of MEG3 inhibited HMGB1 expression in H9c2 cells. In conclusion, targeting MEG3 ameliorates imatinib-induced injury of cardiomyocytes via regulating miR-129-5p/HMGB1 axis.
Copyright © 2023 Peng Tang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
Similar articles
-
MEG3 aggravates hypoxia/reoxygenation induced apoptosis of renal tubular epithelial cells via the miR-129-5p/HMGB1 axis.J Biochem Mol Toxicol. 2021 Feb;35(2):e22649. doi: 10.1002/jbt.22649. Epub 2020 Nov 11. J Biochem Mol Toxicol. 2021. PMID: 33175458
-
LncRNA highly upregulated in liver cancer regulates imatinib resistance in chronic myeloid leukemia via the miR-150-5p/MCL1 axis.Anticancer Drugs. 2021 Apr 1;32(4):427-436. doi: 10.1097/CAD.0000000000001019. Anticancer Drugs. 2021. PMID: 33587348
-
LncRNA OIP5-AS1 facilitates ox-LDL-induced endothelial cell injury through the miR-98-5p/HMGB1 axis.Mol Cell Biochem. 2021 Jan;476(1):443-455. doi: 10.1007/s11010-020-03921-5. Epub 2020 Sep 29. Mol Cell Biochem. 2021. PMID: 32990894
-
Long Noncoding RNA Taurine-Upregulated Gene 1 Knockdown Protects Cardiomyocytes Against Hypoxia/Reoxygenation-induced Injury Through Regulating miR-532-5p/Sox8 Axis.J Cardiovasc Pharmacol. 2020 Nov;76(5):556-563. doi: 10.1097/FJC.0000000000000895. J Cardiovasc Pharmacol. 2020. PMID: 32833900
-
Long noncoding RNA PCA3 regulates prostate cancer through sponging miR-218-5p and modulating high mobility group box 1.J Cell Physiol. 2019 Aug;234(8):13097-13109. doi: 10.1002/jcp.27980. Epub 2018 Dec 19. J Cell Physiol. 2019. Retraction in: J Cell Physiol. 2022 Mar;237(3):2013. doi: 10.1002/jcp.30526. PMID: 30569456 Retracted.
Cited by
-
Deciphering the roles of cellular and extracellular non-coding RNAs in chemotherapy-induced cardiotoxicity.Mol Cell Biochem. 2024 Nov 1. doi: 10.1007/s11010-024-05143-5. Online ahead of print. Mol Cell Biochem. 2024. PMID: 39485641 Review.
References
-
- Castillo-Casas J. M., Caño-Carrillo S., Sánchez-Fernández C., Franco D., Lozano-Velasco E. Comparative analysis of heart regeneration: searching for the key to heal the heart—part II: molecular mechanisms of cardiac regeneration. Journal of Cardiovascular Development and Disease . 2023;10(9) doi: 10.3390/jcdd10090357.357 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

