Modeling the crosstalk between malignant B cells and their microenvironment in B-cell lymphomas: challenges and opportunities
- PMID: 38022603
- PMCID: PMC10652758
- DOI: 10.3389/fimmu.2023.1288110
Modeling the crosstalk between malignant B cells and their microenvironment in B-cell lymphomas: challenges and opportunities
Abstract
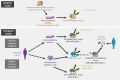
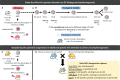
B-cell lymphomas are a group of heterogeneous neoplasms resulting from the clonal expansion of mature B cells arrested at various stages of differentiation. Specifically, two lymphoma subtypes arise from germinal centers (GCs), namely follicular lymphoma (FL) and GC B-cell diffuse large B-cell lymphoma (GCB-DLBCL). In addition to recent advances in describing the genetic landscape of FL and GCB-DLBCL, tumor microenvironment (TME) has progressively emerged as a central determinant of early lymphomagenesis, subclonal evolution, and late progression/transformation. The lymphoma-supportive niche integrates a dynamic and coordinated network of immune and stromal cells defining microarchitecture and mechanical constraints and regulating tumor cell migration, survival, proliferation, and immune escape. Several questions are still unsolved regarding the interplay between lymphoma B cells and their TME, including the mechanisms supporting these bidirectional interactions, the impact of the kinetic and spatial heterogeneity of the tumor niche on B-cell heterogeneity, and how individual genetic alterations can trigger both B-cell intrinsic and B-cell extrinsic signals driving the reprogramming of non-malignant cells. Finally, it is not clear whether these interactions might promote resistance to treatment or, conversely, offer valuable therapeutic opportunities. A major challenge in addressing these questions is the lack of relevant models integrating tumor cells with specific genetic hits, non-malignant cells with adequate functional properties and organization, extracellular matrix, and biomechanical forces. We propose here an overview of the 3D in vitro models, xenograft approaches, and genetically-engineered mouse models recently developed to study GC B-cell lymphomas with a specific focus on the pros and cons of each strategy in understanding B-cell lymphomagenesis and evaluating new therapeutic strategies.
Keywords: 3D models; diffuse large B-cell lymphoma; follicular lymphoma; genetically-engineered mouse models; germinal center; stromal cells; tumor microenvironment; xenografts.
Copyright © 2023 Brauge, Dessauge, Creusat and Tarte.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Human B Lymphomas Reveal Their Secrets Through Genetic Mouse Models.Front Immunol. 2021 Jul 16;12:683597. doi: 10.3389/fimmu.2021.683597. eCollection 2021. Front Immunol. 2021. PMID: 34335584 Free PMC article. Review.
-
Cell cross talk within the lymphoma tumor microenvironment: follicular lymphoma as a paradigm.Blood. 2024 Mar 21;143(12):1080-1090. doi: 10.1182/blood.2023021000. Blood. 2024. PMID: 38096368
-
Neutrophils trigger a NF-κB dependent polarization of tumor-supportive stromal cells in germinal center B-cell lymphomas.Oncotarget. 2015 Jun 30;6(18):16471-87. doi: 10.18632/oncotarget.4106. Oncotarget. 2015. PMID: 26158216 Free PMC article.
-
Microenvironment signaling driving lymphomagenesis.Curr Opin Hematol. 2018 Jul;25(4):335-345. doi: 10.1097/MOH.0000000000000440. Curr Opin Hematol. 2018. PMID: 29746265 Review.
-
Bone Marrow Lymphoid Niche Adaptation to Mature B Cell Neoplasms.Front Immunol. 2021 Dec 8;12:784691. doi: 10.3389/fimmu.2021.784691. eCollection 2021. Front Immunol. 2021. PMID: 34956214 Free PMC article. Review.
Cited by
-
Applications of Multimodal Artificial Intelligence in Non-Hodgkin Lymphoma B Cells.Biomedicines. 2024 Aug 5;12(8):1753. doi: 10.3390/biomedicines12081753. Biomedicines. 2024. PMID: 39200217 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

