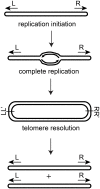
The telomere resolvase, TelA, utilizes an underwound pre-cleavage intermediate to promote hairpin telomere formation
- PMID: 38019799
- PMCID: PMC10686437
- DOI: 10.1371/journal.pone.0294732
The telomere resolvase, TelA, utilizes an underwound pre-cleavage intermediate to promote hairpin telomere formation
Abstract
The telomere resolvase, TelA, forms the hairpin telomeres of the linear chromosome of Agrobacterium tumefaciens in a process referred to as telomere resolution. Telomere resolution is a unique DNA cleavage and rejoining reaction that resolves replicated telomere junctions into a pair of hairpin telomeres. Telomere resolvases utilize a reaction mechanism with similarities to that of topoisomerase-IB enzymes and tyrosine recombinases. The reaction proceeds without the need for high-energy cofactors due to the use of a covalent, enzyme-cleaved DNA intermediate that stores the bond energy of the cleaved bonds in 3'-phosphotyrosyl linkages. The cleaved DNA strands are then refolded into a hairpin conformation and the 5'-OH ends of the refolded strands attack the 3'-phosphotyrosine linkages in order to rejoin the DNA strands into hairpin telomeres. Because this kind of reaction mechanism is, in principle, reversible it is unclear how TelA controls the direction of the reaction and propels the reaction to completion. We present evidence that TelA forms and/or stabilizes a pre-cleavage intermediate that features breakage of the four central basepairs between the scissile phosphates prior to DNA cleavage to help propel the reaction forwards, thus preventing abortive cleavage and rejoining cycles that regenerate the substrate DNA. We identify eight TelA sidechains, located in the hairpin-binding module and catalytic domains of TelA, implicated in this process. These mutants were deficient for telomere resolution on parental replicated telomere junctions but were rescued by introduction of substrate modifications that mimic unwinding of the DNA between the scissile phosphates.
Copyright: © 2023 Balouchi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Spring loading a pre-cleavage intermediate for hairpin telomere formation.Nucleic Acids Res. 2015 Jul 13;43(12):6062-74. doi: 10.1093/nar/gkv497. Epub 2015 May 24. Nucleic Acids Res. 2015. PMID: 26007659 Free PMC article.
-
The N-terminal domain of the Agrobacterium tumefaciens telomere resolvase, TelA, regulates its DNA cleavage and rejoining activities.J Biol Chem. 2022 May;298(5):101951. doi: 10.1016/j.jbc.2022.101951. Epub 2022 Apr 18. J Biol Chem. 2022. PMID: 35447111 Free PMC article.
-
Single stranded DNA annealing is a conserved activity of telomere resolvases.PLoS One. 2021 Feb 4;16(2):e0246212. doi: 10.1371/journal.pone.0246212. eCollection 2021. PLoS One. 2021. PMID: 33539370 Free PMC article.
-
Hairpin Telomere Resolvases.Microbiol Spectr. 2014 Dec;2(6). doi: 10.1128/microbiolspec.MDNA3-0023-2014. Microbiol Spectr. 2014. PMID: 26104454 Review.
-
Split target specificity of ResT: a design for protein delivery, site selectivity and regulation of enzyme activity?Mol Microbiol. 2007 May;64(3):575-9. doi: 10.1111/j.1365-2958.2007.05700.x. Mol Microbiol. 2007. PMID: 17462008 Review.
Cited by
-
Design and characterization of hyperactive mutants of the Agrobacterium tumefaciens telomere resolvase, TelA.PLoS One. 2024 Jul 25;19(7):e0307590. doi: 10.1371/journal.pone.0307590. eCollection 2024. PLoS One. 2024. PMID: 39052566 Free PMC article.
-
Structure analysis of the telomere resolvase from the Lyme disease spirochete Borrelia garinii reveals functional divergence of its C-terminal domain.Nucleic Acids Res. 2024 Aug 12;52(14):8431-8442. doi: 10.1093/nar/gkae580. Nucleic Acids Res. 2024. PMID: 38979576 Free PMC article.
References
-
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al.. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390(6660):580–6. - PubMed
-
- Hertwig S, Klein I, Lurz R, Lanka E, Appel B. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol Microbiol. 2003;48(4):989–1003. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

