Effects of phosphorylation on Drp1 activation by its receptors, actin, and cardiolipin
- PMID: 38019609
- PMCID: PMC10881151
- DOI: 10.1091/mbc.E23-11-0427
Effects of phosphorylation on Drp1 activation by its receptors, actin, and cardiolipin
Abstract
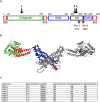
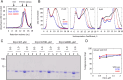
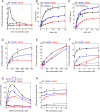
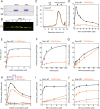
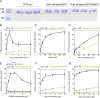
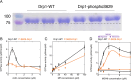
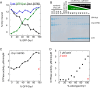
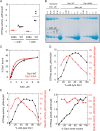
Drp1 is a dynamin family GTPase required for mitochondrial and peroxisomal division. Oligomerization increases Drp1 GTPase activity through interactions between neighboring GTPase domains. In cells, Drp1 is regulated by several factors including Drp1 receptors, actin filaments, cardiolipin, and phosphorylation at two sites: S579 and S600. Commonly, phosphorylation of S579 is considered activating, while S600 phosphorylation is considered inhibiting. However, direct effects of phosphorylation on Drp1 GTPase activity have not been investigated in detail. Here, we compare effects of S579 and S600 phosphorylation on purified Drp1, using phosphomimetic mutants and in vitro phosphorylation. Both phosphomimetic mutants are shifted toward smaller oligomers. Both phosphomimetic mutations maintain basal GTPase activity, but eliminate GTPase stimulation by actin and decrease GTPase stimulation by cardiolipin, Mff, and MiD49. Phosphorylation of S579 by Erk2 produces similar effects. When mixed with wildtype Drp1, both S579D and S600D phosphomimetic mutants reduce the actin-stimulated GTPase activity of Drp1-WT. Conversely, a Drp1 mutant (K38A) lacking GTPase activity stimulates Drp1-WT GTPase activity under both basal and actin-stimulated conditions. These results suggest that the effect of S579 phosphorylation is not to activate Drp1 directly. In addition, our results suggest that nearest neighbor interactions within the Drp1 oligomer affect catalytic activity.
Figures
Update of
-
Effects of phosphorylation on Drp1 activation by its receptors, actin, and cardiolipin.bioRxiv [Preprint]. 2023 Aug 20:2023.08.20.554022. doi: 10.1101/2023.08.20.554022. bioRxiv. 2023. Update in: Mol Biol Cell. 2024 Feb 1;35(2):ar16. doi: 10.1091/mbc.E23-11-0427. PMID: 37645886 Free PMC article. Updated. Preprint.
Similar articles
-
Effects of phosphorylation on Drp1 activation by its receptors, actin, and cardiolipin.bioRxiv [Preprint]. 2023 Aug 20:2023.08.20.554022. doi: 10.1101/2023.08.20.554022. bioRxiv. 2023. Update in: Mol Biol Cell. 2024 Feb 1;35(2):ar16. doi: 10.1091/mbc.E23-11-0427. PMID: 37645886 Free PMC article. Updated. Preprint.
-
Mff oligomerization is required for Drp1 activation and synergy with actin filaments during mitochondrial division.Mol Biol Cell. 2021 Oct 1;32(20):ar5. doi: 10.1091/mbc.E21-04-0224. Epub 2021 Aug 4. Mol Biol Cell. 2021. PMID: 34347505 Free PMC article.
-
Distinct Splice Variants of Dynamin-related Protein 1 Differentially Utilize Mitochondrial Fission Factor as an Effector of Cooperative GTPase Activity.J Biol Chem. 2016 Jan 1;291(1):493-507. doi: 10.1074/jbc.M115.680181. Epub 2015 Nov 17. J Biol Chem. 2016. PMID: 26578513 Free PMC article.
-
The Drp1-Mediated Mitochondrial Fission Protein Interactome as an Emerging Core Player in Mitochondrial Dynamics and Cardiovascular Disease Therapy.Int J Mol Sci. 2023 Mar 17;24(6):5785. doi: 10.3390/ijms24065785. Int J Mol Sci. 2023. PMID: 36982862 Free PMC article. Review.
-
The role of Drp1 adaptor proteins MiD49 and MiD51 in mitochondrial fission: implications for human disease.Clin Sci (Lond). 2016 Nov 1;130(21):1861-74. doi: 10.1042/CS20160030. Clin Sci (Lond). 2016. PMID: 27660309 Review.
Cited by
-
Serine 39 in the GTP-binding domain of Drp1 is involved in shaping mitochondrial morphology.FEBS Open Bio. 2024 Jul;14(7):1147-1165. doi: 10.1002/2211-5463.13820. Epub 2024 May 17. FEBS Open Bio. 2024. PMID: 38760979 Free PMC article.
-
Systemic phospho-defective and phospho-mimetic Drp1 mice exhibit normal growth and development with altered anxiety-like behavior.iScience. 2024 May 3;27(6):109874. doi: 10.1016/j.isci.2024.109874. eCollection 2024 Jun 21. iScience. 2024. PMID: 38784001 Free PMC article.
-
The Role of Cardiolipin in Mitochondrial Function and Neurodegenerative Diseases.Cells. 2024 Mar 30;13(7):609. doi: 10.3390/cells13070609. Cells. 2024. PMID: 38607048 Free PMC article. Review.
References
-
- Abe Y, Wanders RJA, Waterham HR, Mandel H, Falik-Zaccai TC, Ishihara N, Fujiki Y (2023). Genetic defects in peroxisome morphogenesis (Pex11β, dynamin-like protein 1, and nucleoside diphosphate kinase 3) affect docosahexaenoic acid-phospholipid metabolism. J Inherit Metab Dis 46, 273–285. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

