Re-Evaluating Human Cytomegalovirus Vaccine Design: Prediction of T Cell Epitopes
- PMID: 38005961
- PMCID: PMC10674879
- DOI: 10.3390/vaccines11111629
Re-Evaluating Human Cytomegalovirus Vaccine Design: Prediction of T Cell Epitopes
Abstract
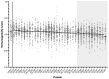
HCMV vaccine development has traditionally focused on viral antigens identified as key targets of neutralizing antibody (NAb) and/or T cell responses in healthy adults with chronic HCMV infection, such as glycoprotein B (gB), the glycoprotein H-anchored pentamer complex (PC), and the unique long 83 (UL83)-encoded phosphoprotein 65 (pp65). However, the protracted absence of a licensed HCMV vaccine that reduces the risk of infection in pregnancy regardless of serostatus warrants a systematic reassessment of assumptions informing vaccine design. To illustrate this imperative, we considered the hypothesis that HCMV proteins infrequently detected as targets of T cell responses may contain important vaccine antigens. Using an extant dataset from a T cell profiling study, we tested whether HCMV proteins recognized by only a small minority of participants encompass any T cell epitopes. Our analyses demonstrate a prominent skewing of T cell responses away from most viral proteins-although they contain robust predicted CD8 T cell epitopes-in favor of a more restricted set of proteins. Our findings raise the possibility that HCMV may benefit from evading the T cell recognition of certain key proteins and that, contrary to current vaccine design approaches, including them as vaccine antigens could effectively take advantage of this vulnerability.
Keywords: bioinformatics; cytomegalovirus; unconventional T cell antigen candidates; vaccine.
Conflict of interest statement
P.A.B. and S.S.I. declare no conflicts of interest. LG received grant funding from Moderna Therapeutics, Inc. (Cambridge, MA, USA).
Figures
Similar articles
-
Vaccine-Derived Neutralizing Antibodies to the Human Cytomegalovirus gH/gL Pentamer Potently Block Primary Cytotrophoblast Infection.J Virol. 2015 Dec;89(23):11884-98. doi: 10.1128/JVI.01701-15. Epub 2015 Sep 16. J Virol. 2015. PMID: 26378171 Free PMC article.
-
Human cytomegalovirus (HCMV) infection/re-infection: development of a protective HCMV vaccine.New Microbiol. 2019 Jan;42(1):1-20. Epub 2019 Jan 21. New Microbiol. 2019. PMID: 30671581 Review.
-
Human cytomegalovirus proteins pp65 and immediate early protein 1 are common targets for CD8+ T cell responses in children with congenital or postnatal human cytomegalovirus infection.J Immunol. 2004 Feb 15;172(4):2256-64. doi: 10.4049/jimmunol.172.4.2256. J Immunol. 2004. PMID: 14764694
-
Dense Bodies of a gH/gL/UL128/UL130/UL131 Pentamer-Repaired Towne Strain of Human Cytomegalovirus Induce an Enhanced Neutralizing Antibody Response.J Virol. 2019 Aug 13;93(17):e00931-19. doi: 10.1128/JVI.00931-19. Print 2019 Sep 1. J Virol. 2019. PMID: 31189713 Free PMC article.
-
The next generation recombinant human cytomegalovirus vaccine candidates-beyond gB.Vaccine. 2012 Nov 19;30(49):6980-90. doi: 10.1016/j.vaccine.2012.09.056. Epub 2012 Oct 3. Vaccine. 2012. PMID: 23041121 Review.
References
-
- [(accessed on 2 May 2023)]. Available online: https://ourworldindata.org/grapher/births-and-deaths-projected-to-2100.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

