MiR-155 Negatively Regulates Anti-Viral Innate Responses among HIV-Infected Progressors
- PMID: 38005883
- PMCID: PMC10675553
- DOI: 10.3390/v15112206
MiR-155 Negatively Regulates Anti-Viral Innate Responses among HIV-Infected Progressors
Abstract
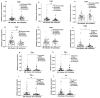
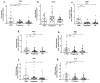
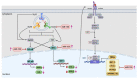
HIV infection impairs host immunity, leading to progressive disease. An anti-retroviral treatment efficiently controls viremia but cannot completely restore the immune dysfunction in HIV-infected individuals. Both host and viral factors determine the rate of disease progression. Among the host factors, innate immunity plays a critical role; however, the mechanism(s) associated with dysfunctional innate responses are poorly understood among HIV disease progressors, which was investigated here. The gene expression profiles of TLRs and innate cytokines in HIV-infected (LTNPs and progressors) and HIV-uninfected individuals were examined. Since the progressors showed a dysregulated TLR-mediated innate response, we investigated the role of TLR agonists in restoring the innate functions of the progressors. The stimulation of PBMCs with TLR3 agonist-poly:(I:C), TLR7 agonist-GS-9620 and TLR9 agonist-ODN 2216 resulted in an increased expression of IFN-α, IFN-β and IL-6. Interestingly, the expression of IFITM3, BST-2, IFITM-3, IFI-16 was also increased upon stimulation with TLR3 and TLR7 agonists, respectively. To further understand the molecular mechanism involved, the role of miR-155 was explored. Increased miR-155 expression was noted among the progressors. MiR-155 inhibition upregulated the expression of TLR3, NF-κB, IRF-3, TNF-α and the APOBEC-3G, IFITM-3, IFI-16 and BST-2 genes in the PBMCs of the progressors. To conclude, miR-155 negatively regulates TLR-mediated cytokines as wel l as the expression of host restriction factors, which play an important role in mounting anti-HIV responses; hence, targeting miR-155 might be helpful in devising strategic approaches towards alleviating HIV disease progression.
Keywords: HIV; LTNPs; TLRs; disease progression; host restriction factors; innate; miR-155.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
TLR activation pathways in HIV-1-exposed seronegative individuals.J Immunol. 2010 Mar 1;184(5):2710-7. doi: 10.4049/jimmunol.0902463. Epub 2010 Feb 1. J Immunol. 2010. PMID: 20124101
-
TLR7/TLR8 Activation Restores Defective Cytokine Secretion by Myeloid Dendritic Cells but Not by Plasmacytoid Dendritic Cells in HIV-Infected Pregnant Women and Newborns.PLoS One. 2013 Jun 27;8(6):e67036. doi: 10.1371/journal.pone.0067036. Print 2013. PLoS One. 2013. PMID: 23826189 Free PMC article.
-
A role for microRNA-155 modulation in the anti-HIV-1 effects of Toll-like receptor 3 stimulation in macrophages.PLoS Pathog. 2012 Sep;8(9):e1002937. doi: 10.1371/journal.ppat.1002937. Epub 2012 Sep 20. PLoS Pathog. 2012. PMID: 23028330 Free PMC article.
-
The Role of Innate Immunity in Natural Elite Controllers of HIV-1 Infection.Front Immunol. 2022 Feb 8;13:780922. doi: 10.3389/fimmu.2022.780922. eCollection 2022. Front Immunol. 2022. PMID: 35211115 Free PMC article. Review.
-
Cross-talk between virus and host innate immunity in pediatric HIV-1 infection and disease progression.New Microbiol. 2012 Jul;35(3):249-57. Epub 2012 Jun 30. New Microbiol. 2012. PMID: 22842595 Review.
Cited by
-
Role of microRNAs in Immune Regulation with Translational and Clinical Applications.Int J Mol Sci. 2024 Feb 5;25(3):1942. doi: 10.3390/ijms25031942. Int J Mol Sci. 2024. PMID: 38339220 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

