Covalent Inhibitors from Saudi Medicinal Plants Target RNA-Dependent RNA Polymerase (RdRp) of SARS-CoV-2
- PMID: 38005857
- PMCID: PMC10675690
- DOI: 10.3390/v15112175
Covalent Inhibitors from Saudi Medicinal Plants Target RNA-Dependent RNA Polymerase (RdRp) of SARS-CoV-2
Abstract
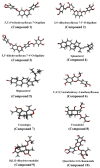
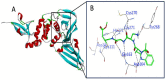
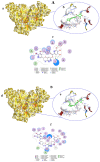
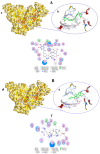
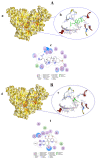
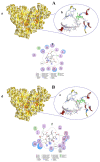
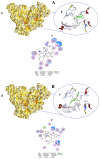
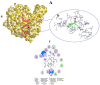
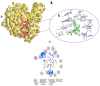
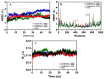
COVID-19, a disease caused by SARS-CoV-2, has caused a huge loss of human life, and the number of deaths is still continuing. Despite the lack of repurposed drugs and vaccines, the search for potential small molecules to inhibit SARS-CoV-2 is in demand. Hence, we relied on the drug-like characters of ten phytochemicals (compounds 1-10) that were previously isolated and purified by our research team from Saudi medicinal plants. We computationally evaluated the inhibition of RNA-dependent RNA polymerase (RdRp) by compounds 1-10. Non-covalent (reversible) docking of compounds 1-10 with RdRp led to the formation of a hydrogen bond with template primer nucleotides (A and U) and key amino acid residues (ASP623, LYS545, ARG555, ASN691, SER682, and ARG553) in its active pocket. Covalent (irreversible) docking revealed that compounds 7, 8, and 9 exhibited their irreversible nature of binding with CYS813, a crucial amino acid in the palm domain of RdRP. Molecular dynamic (MD) simulation analysis by RMSD, RMSF, and Rg parameters affirmed that RdRP complexes with compounds 7, 8, and 9 were stable and showed less deviation. Our data provide novel information on compounds 7, 8, and 9 that demonstrated their non-nucleoside and irreversible interaction capabilities to inhibit RdRp and shed new scaffolds as antivirals against SARS-CoV-2.
Keywords: COVID-19; MD simulation; RdRp; SARS-CoV-2; docking; medicinal plants; phytochemicals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Identifying non-nucleoside inhibitors of RNA-dependent RNA-polymerase of SARS-CoV-2 through per-residue energy decomposition-based pharmacophore modeling, molecular docking, and molecular dynamics simulation.J Infect Public Health. 2023 Apr;16(4):501-519. doi: 10.1016/j.jiph.2023.02.009. Epub 2023 Feb 14. J Infect Public Health. 2023. PMID: 36801630 Free PMC article.
-
Investigating the potential of natural compounds as novel inhibitors of SARS-CoV-2 RdRP using computational approaches.Biotechnol Genet Eng Rev. 2024 Nov;40(3):1535-1555. doi: 10.1080/02648725.2023.2195240. Epub 2023 Mar 30. Biotechnol Genet Eng Rev. 2024. PMID: 36994810 Review.
-
Alkaloids and flavonoids from African phytochemicals as potential inhibitors of SARS-Cov-2 RNA-dependent RNA polymerase: an in silico perspective.Antivir Chem Chemother. 2020 Jan-Dec;28:2040206620984076. doi: 10.1177/2040206620984076. Antivir Chem Chemother. 2020. PMID: 33372806 Free PMC article.
-
Identification of Phytochemicals from Arabian Peninsula Medicinal Plants as Strong Binders to SARS-CoV-2 Proteases (3CLPro and PLPro) by Molecular Docking and Dynamic Simulation Studies.Molecules. 2024 Feb 25;29(5):998. doi: 10.3390/molecules29050998. Molecules. 2024. PMID: 38474509 Free PMC article.
-
Potential Candidates against COVID-19 Targeting RNA-Dependent RNA Polymerase: A Comprehensive Review.Curr Pharm Biotechnol. 2022;23(3):396-419. doi: 10.2174/1389201022666210421102513. Curr Pharm Biotechnol. 2022. PMID: 33882805 Review.
References
-
- Bi Q., Wu Y., Mei S., Ye C., Zou X., Zhang Z., Liu X., Wei L., Truelove S.A., Zhang T., et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: A retrospective cohort study. Lancet Infect. Dis. 2020;20:911–919. doi: 10.1016/S1473-3099(20)30287-5. - DOI - PMC - PubMed
-
- WHO WHO Coronavirus (COVID-19) Dashboard. [(accessed on 9 October 2023)];2023 Available online: https://covid19.who.int.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

