Evaluation of the Neutralizing Antibody STE90-C11 against SARS-CoV-2 Delta Infection and Its Recognition of Other Variants of Concerns
- PMID: 38005829
- PMCID: PMC10675157
- DOI: 10.3390/v15112153
Evaluation of the Neutralizing Antibody STE90-C11 against SARS-CoV-2 Delta Infection and Its Recognition of Other Variants of Concerns
Abstract
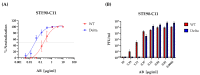
As of now, the COVID-19 pandemic has spread to over 770 million confirmed cases and caused approximately 7 million deaths. While several vaccines and monoclonal antibodies (mAb) have been developed and deployed, natural selection against immune recognition of viral antigens by antibodies has fueled the evolution of new emerging variants and limited the immune protection by vaccines and mAb. To optimize the efficiency of mAb, it is imperative to understand how they neutralize the variants of concern (VoCs) and to investigate the mutations responsible for immune escape. In this study, we show the in vitro neutralizing effects of a previously described monoclonal antibody (STE90-C11) against the SARS-CoV-2 Delta variant (B.1.617.2) and its in vivo effects in therapeutic and prophylactic settings. We also show that the Omicron variant avoids recognition by this mAb. To define which mutations are responsible for the escape in the Omicron variant, we used a library of pseudovirus mutants carrying each of the mutations present in the Omicron VoC individually. We show that either 501Y or 417K point mutations were sufficient for the escape of Omicron recognition by STE90-C11. To test how escape mutations act against a combination of antibodies, we tested the same library against bispecific antibodies, recognizing two discrete regions of the spike antigen. While Omicron escaped the control by the bispecific antibodies, the same antibodies controlled all mutants with individual mutations.
Keywords: Delta variant; SARS-CoV-2; bispecific antibodies; intranasal administration; intravenous administration; mice experiments; monoclonal antibody; pseudovirus assay; single mutations.
Conflict of interest statement
The authors declare the following conflict of interest. L.Č.-Š. was a consultant to CORAT Therapeutics GmbH, a subsidiary of YUMAB GmbH. T.S., A.F., S.D. and M.H. are founders and shareholders of YUMAB GmbH. F.B., M.S., P.A.H., S.D. and M.H. are inventors on a patent application on blocking antibodies against SARS-CoV-2.
Figures



Similar articles
-
A Bispecific Antibody Targeting RBD and S2 Potently Neutralizes SARS-CoV-2 Omicron and Other Variants of Concern.J Virol. 2022 Aug 24;96(16):e0077522. doi: 10.1128/jvi.00775-22. Epub 2022 Aug 2. J Virol. 2022. PMID: 35916510 Free PMC article.
-
Shared N417-Dependent Epitope on the SARS-CoV-2 Omicron, Beta, and Delta Plus Variants.J Virol. 2022 Aug 10;96(15):e0055822. doi: 10.1128/jvi.00558-22. Epub 2022 Jul 13. J Virol. 2022. PMID: 35867572 Free PMC article.
-
SARS-CoV-2 evolves to reduce but not abolish neutralizing action.J Med Virol. 2023 Jan;95(1):e28207. doi: 10.1002/jmv.28207. Epub 2022 Oct 19. J Med Virol. 2023. PMID: 36217880 Free PMC article.
-
Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission.Rev Med Virol. 2022 Sep;32(5):e2381. doi: 10.1002/rmv.2381. Epub 2022 Jul 20. Rev Med Virol. 2022. PMID: 35856385 Free PMC article. Review.
-
Clinical virology and effect of Covid-19 vaccination and monoclonal antibodies against highly infectious SARS- CoV-2 omicron sub variant BF.7 (BA.5.2.1.7): A systematic review.Virology. 2023 Jul;584:38-43. doi: 10.1016/j.virol.2023.04.007. Epub 2023 May 19. Virology. 2023. PMID: 37229914 Free PMC article. Review.
Cited by
-
Systematical assessment of the impact of single spike mutations of SARS-CoV-2 Omicron sub-variants on the neutralization capacity of post-vaccination sera.Front Immunol. 2023 Nov 10;14:1288794. doi: 10.3389/fimmu.2023.1288794. eCollection 2023. Front Immunol. 2023. PMID: 38022629 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
- COFONI 10ff22/Niedersächsisches Ministerium für Wissenschaft und Kultur
- 14-76103-184 CORONA-2/20/Niedersächsisches Ministerium für Wissenschaft und Kultur
- PIE-008/Helmholtz Association of German Research Centres
- KA1-Co-02/Helmholtz Association of German Research Centres
- Horizon2020, 101003650/European Union
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

