C8ORF88: A Novel eIF4E-Binding Protein
- PMID: 38003019
- PMCID: PMC10670996
- DOI: 10.3390/genes14112076
C8ORF88: A Novel eIF4E-Binding Protein
Abstract
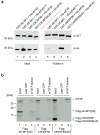
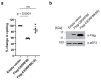
Translation initiation in eukaryotes is regulated at several steps, one of which involves the availability of the cap binding protein to participate in cap-dependent protein synthesis. Binding of eIF4E to translational repressors (eIF4E-binding proteins [4E-BPs]) suppresses translation and is used by cells to link extra- and intracellular cues to protein synthetic rates. The best studied of these interactions involves repression of translation by 4E-BP1 upon inhibition of the PI3K/mTOR signaling pathway. Herein, we characterize a novel 4E-BP, C8ORF88, whose expression is predominantly restricted to early spermatids. C8ORF88:eIF4E interaction is dependent on the canonical eIF4E binding motif (4E-BM) present in other 4E-BPs. Whereas 4E-BP1:eIF4E interaction is dependent on the phosphorylation of 4E-BP1, these sites are not conserved in C8ORF88 indicating a different mode of regulation.
Keywords: C8ORF88; cap-dependent translation; eIF4E; gene expression; mRNA; protein synthesis; translation initiation.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. A.Y. was employed by Arcalis Inc. at time of publication.
Figures



Similar articles
-
Caspase cleavage of initiation factor 4E-binding protein 1 yields a dominant inhibitor of cap-dependent translation and reveals a novel regulatory motif.Mol Cell Biol. 2002 Mar;22(6):1674-83. doi: 10.1128/MCB.22.6.1674-1683.2002. Mol Cell Biol. 2002. PMID: 11865047 Free PMC article.
-
Mitosis-related phosphorylation of the eukaryotic translation suppressor 4E-BP1 and its interaction with eukaryotic translation initiation factor 4E (eIF4E).J Biol Chem. 2019 Aug 2;294(31):11840-11852. doi: 10.1074/jbc.RA119.008512. Epub 2019 Jun 14. J Biol Chem. 2019. PMID: 31201269 Free PMC article.
-
Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1.J Virol. 2002 Oct;76(20):10177-87. doi: 10.1128/jvi.76.20.10177-10187.2002. J Virol. 2002. PMID: 12239292 Free PMC article.
-
Control of the eIF4E activity: structural insights and pharmacological implications.Cell Mol Life Sci. 2021 Nov;78(21-22):6869-6885. doi: 10.1007/s00018-021-03938-z. Epub 2021 Sep 19. Cell Mol Life Sci. 2021. PMID: 34541613 Free PMC article. Review.
-
4E-BP1, a multifactor regulated multifunctional protein.Cell Cycle. 2016;15(6):781-6. doi: 10.1080/15384101.2016.1151581. Cell Cycle. 2016. PMID: 26901143 Free PMC article. Review.
References
-
- Rhoads R.E., Joshi-Barve S., Rinker-Schaeffer C. Mechanism of action and regulation of protein synthesis initiation factor 4E: Effects on mRNA discrimination, cellular growth rate, and oncogenesis. Prog. Nucleic Acid Res. Mol. Biol. 1993;46:183–219. doi: 10.1016/s0079-6603(08)61022-3. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

