The Novel Link between Gene Expression Profiles of Adult T-Cell Leukemia/Lymphoma Patients' Peripheral Blood Lymphocytes and Ferroptosis Susceptibility
- PMID: 38002949
- PMCID: PMC10671613
- DOI: 10.3390/genes14112005
The Novel Link between Gene Expression Profiles of Adult T-Cell Leukemia/Lymphoma Patients' Peripheral Blood Lymphocytes and Ferroptosis Susceptibility
Abstract
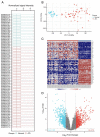
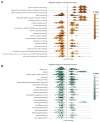
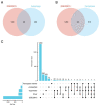
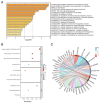
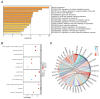
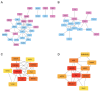
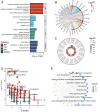
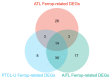
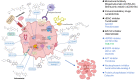
Ferroptosis, a regulated cell death dependent on iron, has garnered attention as a potential broad-spectrum anticancer approach in leukemia research. However, there has been limited ferroptosis research on ATL, an aggressive T-cell malignancy caused by HTLV-1 infection. Our study employs bioinformatic analysis, utilizing dataset GSE33615, to identify 46 ferroptosis-related DEGs and 26 autophagy-related DEGs in ATL cells. These DEGs are associated with various cellular responses, chemical stress, and iron-related pathways. Autophagy-related DEGs are linked to autophagy, apoptosis, NOD-like receptor signaling, TNF signaling, and the insulin resistance pathway. PPI network analysis revealed 10 hub genes and related biomolecules. Moreover, we predicted crucial miRNAs, transcription factors, and potential pharmacological compounds. We also screened the top 20 medications based on upregulated DEGs. In summary, our study establishes an innovative link between ATL treatment and ferroptosis, offering promising avenues for novel therapeutic strategies in ATL.
Keywords: adult T-cell leukemia/lymphoma (ATL); bioinformatic analysis; ferroptosis; precision medicine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Silencing LCN2 enhances RSL3-induced ferroptosis in T cell acute lymphoblastic leukemia.Gene. 2023 Aug 30;879:147597. doi: 10.1016/j.gene.2023.147597. Epub 2023 Jun 28. Gene. 2023. PMID: 37390872
-
Decreased RORC expression and downstream signaling in HTLV-1-associated adult T-cell lymphoma/leukemia uncovers an antiproliferative IL17 link: A potential target for immunotherapy?Int J Cancer. 2019 Apr 1;144(7):1664-1675. doi: 10.1002/ijc.31922. Epub 2018 Dec 18. Int J Cancer. 2019. PMID: 30303535 Free PMC article.
-
YM155 suppresses cell proliferation and induces cell death in human adult T-cell leukemia/lymphoma cells.Leuk Res. 2015 Dec;39(12):1473-9. doi: 10.1016/j.leukres.2015.10.012. Epub 2015 Oct 27. Leuk Res. 2015. PMID: 26547260
-
[New treatment strategy for adult T-cell leukemia targeting for anti-tumor immunity and a longevity gene-encoded protein].Yakugaku Zasshi. 2011;131(7):1061-72. doi: 10.1248/yakushi.131.1061. Yakugaku Zasshi. 2011. PMID: 21720136 Review. Japanese.
-
Viral, genetic, and immune factors in the oncogenesis of adult T-cell leukemia/lymphoma.Int J Hematol. 2023 Apr;117(4):504-511. doi: 10.1007/s12185-023-03547-5. Epub 2023 Jan 27. Int J Hematol. 2023. PMID: 36705848 Review.
Cited by
-
Key candidate genes and pathways in T lymphoblastic leukemia/lymphoma identified by bioinformatics and serological analyses.Front Immunol. 2024 Feb 23;15:1341255. doi: 10.3389/fimmu.2024.1341255. eCollection 2024. Front Immunol. 2024. PMID: 38464517 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

