Evaluation of genetic risk of apparently balanced chromosomal rearrangement carriers by breakpoint characterization
- PMID: 37993578
- PMCID: PMC10789712
- DOI: 10.1007/s10815-023-02986-7
Evaluation of genetic risk of apparently balanced chromosomal rearrangement carriers by breakpoint characterization
Abstract
Purpose: To report genetic characteristics and associated risk of chromosomal breaks due to chromosomal rearrangements in large samples.
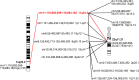
Methods: MicroSeq, a technique that combines chromosome microdissection and next-generation sequencing, was used to identify chromosomal breakpoints. Long-range PCR and Sanger sequencing were used to precisely characterize 100 breakpoints in 50 ABCR carriers.
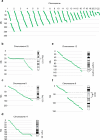
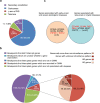
Results: In addition to the recurrent regions of balanced rearrangement breaks in 8q24.13, 11q11.23, and 22q11.21 that had been documented, we have discovered a 10-Mb region of 12q24.13-q24.3 that could potentially be a sparse region of balanced rearrangement breaks. We found that 898 breakpoints caused gene disruption and a total of 188 breakpoints interrupted genes recorded in OMIM. The percentage of breakpoints that disrupted autosomal dominant genes recorded in OMIM was 25.53% (48/188). Fifty-four of the precisely characterized breakpoints had 1-8-bp microhomologous sequences.
Conclusion: Our findings provide a reference for the evaluation of the pathogenicity of mutations in related genes that cause protein truncation in clinical practice. According to the characteristics of breakpoints, non-homologous end joining and microhomology-mediated break-induced replication may be the main mechanism for ABCRs formation.
Keywords: Breakpoint; Chromosome rearrangement; Genetic risk; MicroSeq.
© 2023. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations.J Med Genet. 2013 Mar;50(3):144-50. doi: 10.1136/jmedgenet-2012-101351. Epub 2013 Jan 12. J Med Genet. 2013. PMID: 23315544
-
Whole genome paired-end sequencing elucidates functional and phenotypic consequences of balanced chromosomal rearrangement in patients with developmental disorders.J Med Genet. 2019 Aug;56(8):526-535. doi: 10.1136/jmedgenet-2018-105778. Epub 2019 Mar 28. J Med Genet. 2019. PMID: 30923172
-
Breakpoint mapping at nucleotide resolution in X-autosome balanced translocations associated with clinical phenotypes.Eur J Hum Genet. 2019 May;27(5):760-771. doi: 10.1038/s41431-019-0341-5. Epub 2019 Jan 30. Eur J Hum Genet. 2019. PMID: 30700833 Free PMC article.
-
Characterization of renal cell carcinoma-associated constitutional chromosome abnormalities by genome sequencing.Genes Chromosomes Cancer. 2020 Jun;59(6):333-347. doi: 10.1002/gcc.22833. Epub 2020 Feb 5. Genes Chromosomes Cancer. 2020. PMID: 31943436 Free PMC article. Review.
-
Translocation breakpoints of chromosome 3 in male carriers: a report of twelve cases and a review of the literature.Turk J Med Sci. 2018 Feb 23;48(1):150-156. doi: 10.3906/sag-1704-1. Turk J Med Sci. 2018. PMID: 29479975 Review.
Cited by
-
Characterization of cryptic complex chromosome rearrangements in balanced chromosomal rearrangement carriers and their PGT-SR clinical outcome assessments.Sci Rep. 2024 Sep 5;14(1):20705. doi: 10.1038/s41598-024-70566-5. Sci Rep. 2024. PMID: 39237551 Free PMC article.
References
-
- David D, et al. Comprehensive clinically oriented workflow for nucleotide level resolution and interpretation in prenatal diagnosis of de novo apparently balanced chromosomal translocations in their genomic landscape. Hum Genet. 2020;139:531–543. doi: 10.1007/s00439-020-02121-x. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

