HIV-1 Gag co-localizes with euchromatin histone marks at the nuclear periphery
- PMID: 37991367
- PMCID: PMC10734548
- DOI: 10.1128/jvi.01179-23
HIV-1 Gag co-localizes with euchromatin histone marks at the nuclear periphery
Abstract
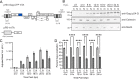
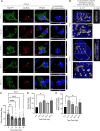
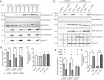
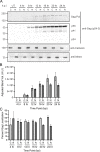
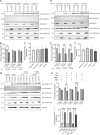
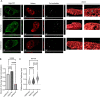
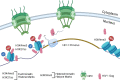
The traditional view of retrovirus assembly posits that packaging of gRNA by HIV-1 Gag occurs in the cytoplasm or at the plasma membrane. However, our previous studies showing that HIV-1 Gag enters the nucleus and binds to USvRNA at transcription sites suggest that gRNA selection may occur in the nucleus. In the present study, we observed that HIV-1 Gag trafficked to the nucleus and co-localized with USvRNA within 8 hours of expression. In infected T cells (J-Lat 10.6) reactivated from latency and in a HeLa cell line stably expressing an inducible Rev-dependent HIV-1 construct, we found that Gag preferentially localized with euchromatin histone marks associated with enhancer and promoter regions near the nuclear periphery, which is the favored site HIV-1 integration. These observations support the innovative hypothesis that HIV-1 Gag associates with euchromatin-associated histones to localize to active transcription sites, promoting capture of newly synthesized gRNA for packaging.
Keywords: HIV-1 Gag; HIV-1 latency reversal; euchromatin localization; nuclear trafficking; retrovirus assembly; subcellular fractionation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Update of
-
HIV-1 Gag colocalizes with euchromatin histone marks at the nuclear periphery.bioRxiv [Preprint]. 2023 Feb 25:2023.02.24.529990. doi: 10.1101/2023.02.24.529990. bioRxiv. 2023. Update in: J Virol. 2023 Dec 21;97(12):e0117923. doi: 10.1128/jvi.01179-23. PMID: 36865288 Free PMC article. Updated. Preprint.
Similar articles
-
HIV-1 Gag colocalizes with euchromatin histone marks at the nuclear periphery.bioRxiv [Preprint]. 2023 Feb 25:2023.02.24.529990. doi: 10.1101/2023.02.24.529990. bioRxiv. 2023. Update in: J Virol. 2023 Dec 21;97(12):e0117923. doi: 10.1128/jvi.01179-23. PMID: 36865288 Free PMC article. Updated. Preprint.
-
HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites.Viruses. 2020 Nov 9;12(11):1281. doi: 10.3390/v12111281. Viruses. 2020. PMID: 33182496 Free PMC article.
-
Visualizing Association of the Retroviral Gag Protein with Unspliced Viral RNA in the Nucleus.mBio. 2020 Apr 7;11(2):e00524-20. doi: 10.1128/mBio.00524-20. mBio. 2020. PMID: 32265329 Free PMC article.
-
The Life-Cycle of the HIV-1 Gag-RNA Complex.Viruses. 2016 Sep 10;8(9):248. doi: 10.3390/v8090248. Viruses. 2016. PMID: 27626439 Free PMC article. Review.
-
Post-Translational Modifications of Retroviral HIV-1 Gag Precursors: An Overview of Their Biological Role.Int J Mol Sci. 2021 Mar 11;22(6):2871. doi: 10.3390/ijms22062871. Int J Mol Sci. 2021. PMID: 33799890 Free PMC article. Review.
Cited by
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.bioRxiv [Preprint]. 2024 Mar 6:2024.01.18.575255. doi: 10.1101/2024.01.18.575255. bioRxiv. 2024. Update in: Retrovirology. 2024 Jun 19;21(1):13. doi: 10.1186/s12977-024-00645-y. PMID: 38293010 Free PMC article. Updated. Preprint.
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.Retrovirology. 2024 Jun 19;21(1):13. doi: 10.1186/s12977-024-00645-y. Retrovirology. 2024. PMID: 38898526 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

