Prediction and validation of common targets in atherosclerosis and non-small cell lung cancer influenced by atorvastatin
- PMID: 37978381
- PMCID: PMC10657002
- DOI: 10.1186/s12906-023-04255-7
Prediction and validation of common targets in atherosclerosis and non-small cell lung cancer influenced by atorvastatin
Abstract
Background: Cardiovascular disease and cancer are the main causes of morbidity and mortality worldwide. Studies have shown that these two diseases may have some common risk factors. Atorvastatin is mainly used for the treatment of atherosclerosis in clinic. A large number of studies show that atorvastatin may produce anti-tumor activities. This study aimed to predict the common targets of atorvastatin against atherosclerosis and non-small cell lung cancer (NSCLC) based on network pharmacology.
Methods: The target genes of atherosclerosis and NSCLC were obtained from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. The disease-target-component model map and the core network were obtained using Cytoscape 3.7.1. The MTS and wound healing assay were used to detect the effect of atorvastatin on cell viability and migration of A549 cells. The expression of potential common target genes of atorvastatin against atherosclerosis and NSCLC were confirmed in A549 cells and lung cancer tissues of patients.
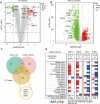
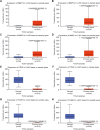
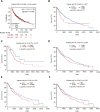
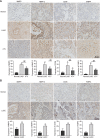
Results: We identified 15 identical pathogenic genes, and four of which (MMP9, MMP12, CD36, and FABP4) were considered as the key target genes of atorvastatin in anti-atherosclerosis and NSCLC. The MTS and wound healing assays revealed that atorvastatin decreased A549 cells migration significantly. Atorvastatin markedly decreased the expression of MMP9, MMP12, CD36, and FABP4 in A549 cells and patients were treated with atorvastatin.
Conclusions: This study demonstrated 15 common pathogenic genes in both atherosclerosis and NSCLC. And verified that MMP 9, MMP 12, CD 36 and FABP 4 might be the common target genes of atorvastatin in anti-atherosclerosis and NSCLC.
Keywords: Atherosclerosis; Atorvastatin; Migration; Network pharmacology; Non-small cell lung cancer.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Inhibition effect of oxyepiberberine isolated from Coptis chinensis franch. On non-small cell lung cancer based on a network pharmacology approach and experimental validation.J Ethnopharmacol. 2021 Oct 5;278:114267. doi: 10.1016/j.jep.2021.114267. Epub 2021 Jun 1. J Ethnopharmacol. 2021. PMID: 34087401
-
Uncovering the anti-NSCLC effects and mechanisms of gypenosides by metabolomics and network pharmacology analysis.J Ethnopharmacol. 2021 Dec 5;281:114506. doi: 10.1016/j.jep.2021.114506. Epub 2021 Aug 6. J Ethnopharmacol. 2021. PMID: 34371113
-
TPX2 Promotes Metastasis and Serves as a Marker of Poor Prognosis in Non-Small Cell Lung Cancer.Med Sci Monit. 2020 Aug 4;26:e925147. doi: 10.12659/MSM.925147. Med Sci Monit. 2020. PMID: 32748897 Free PMC article.
-
Expression Signature and Role of miR-30d-5p in Non-Small Cell Lung Cancer: a Comprehensive Study Based on in Silico Analysis of Public Databases and in Vitro Experiments.Cell Physiol Biochem. 2018;50(5):1964-1987. doi: 10.1159/000494875. Epub 2018 Nov 5. Cell Physiol Biochem. 2018. PMID: 30396166
-
[Blaps rynchopetera affects proliferation, migration, and invasion of non-small cell lung cancer: a study based on network pharmacology and in vivo and in vitro experiments].Zhongguo Zhong Yao Za Zhi. 2023 Jul;48(13):3576-3588. doi: 10.19540/j.cnki.cjcmm.20230202.702. Zhongguo Zhong Yao Za Zhi. 2023. PMID: 37474991 Chinese.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

