rDNA magnification is a unique feature of germline stem cells
- PMID: 37967216
- PMCID: PMC10666004
- DOI: 10.1073/pnas.2314440120
rDNA magnification is a unique feature of germline stem cells
Abstract
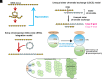
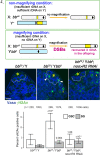
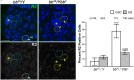
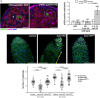
Ribosomal DNA (rDNA) encodes ribosomal RNA and exists as tandem repeats of hundreds of copies in the eukaryotic genome to meet the high demand of ribosome biogenesis. Tandemly repeated DNA elements are inherently unstable; thus, mechanisms must exist to maintain rDNA copy number (CN), in particular in the germline that continues through generations. A phenomenon called rDNA magnification was discovered over 50 y ago in Drosophila as a process that recovers the rDNA CN on chromosomes that harbor minimal CN. Our recent studies indicated that rDNA magnification is the mechanism to maintain rDNA CN under physiological conditions to counteract spontaneous CN loss that occurs during aging. Our previous studies that explored the mechanism of rDNA magnification implied that asymmetric division of germline stem cells (GSCs) may be particularly suited to achieve rDNA magnification. However, it remains elusive whether GSCs are the unique cell type that undergoes rDNA magnification or differentiating germ cells are also capable of magnification. In this study, we provide empirical evidence that suggests that rDNA magnification operates uniquely in GSCs, but not in differentiating germ cells. We further provide computer simulation that suggests that rDNA magnification is only achievable through asymmetric GSC divisions. We propose that despite known plasticity and transcriptomic similarity between GSCs and differentiating germ cells, GSCs' unique ability to divide asymmetrically serves a critical role of maintaining rDNA CN through generations, supporting germline immortality.
Keywords: Drosophila germline; germline immortality; rDNA copy number maintenance; ribosomal DNA.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
Nonrandom sister chromatid segregation mediates rDNA copy number maintenance in Drosophila.Sci Adv. 2022 Jul 29;8(30):eabo4443. doi: 10.1126/sciadv.abo4443. Epub 2022 Jul 27. Sci Adv. 2022. PMID: 35895823 Free PMC article.
-
Transgenerational dynamics of rDNA copy number in Drosophila male germline stem cells.Elife. 2018 Feb 13;7:e32421. doi: 10.7554/eLife.32421. Elife. 2018. PMID: 29436367 Free PMC article.
-
Chromatin and gene expression changes during female Drosophila germline stem cell development illuminate the biology of highly potent stem cells.Elife. 2023 Oct 13;12:RP90509. doi: 10.7554/eLife.90509. Elife. 2023. PMID: 37831064 Free PMC article.
-
Under the magnifying glass: The ups and downs of rDNA copy number.Semin Cell Dev Biol. 2023 Feb 28;136:38-48. doi: 10.1016/j.semcdb.2022.05.006. Epub 2022 May 18. Semin Cell Dev Biol. 2023. PMID: 35595601 Free PMC article. Review.
-
Asymmetric distribution of histones during Drosophila male germline stem cell asymmetric divisions.Chromosome Res. 2013 May;21(3):255-69. doi: 10.1007/s10577-013-9356-x. Chromosome Res. 2013. PMID: 23681658 Free PMC article. Review.
Cited by
-
Transposon and Transgene Tribulations in Mosquitoes: A Perspective of piRNA Proportions.DNA (Basel). 2024 Jun;4(2):104-128. doi: 10.3390/dna4020006. Epub 2024 Mar 30. DNA (Basel). 2024. PMID: 39076684 Free PMC article.
-
RNA polymerase II-mediated rDNA transcription mediates rDNA copy number expansion in Drosophila.PLoS Genet. 2024 May 17;20(5):e1011136. doi: 10.1371/journal.pgen.1011136. eCollection 2024 May. PLoS Genet. 2024. PMID: 38758955 Free PMC article.
References
-
- Kobayashi T., Strategies to maintain the stability of the ribosomal RNA gene repeats–collaboration of recombination, cohesion, and condensation. Genes. Genet. Syst. 81, 155–161 (2006). - PubMed
-
- Henderson A. S., Warburton D., Atwood K. C., Letter: Ribosomal DNA connectives between human acrocentric chromosomes. Nature 245, 95–97 (1973). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

