A closed translocation channel in the substrate-free AAA+ ClpXP protease diminishes rogue degradation
- PMID: 37949857
- PMCID: PMC10638403
- DOI: 10.1038/s41467-023-43145-x
A closed translocation channel in the substrate-free AAA+ ClpXP protease diminishes rogue degradation
Abstract
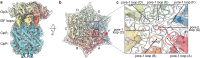
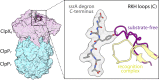
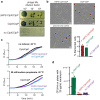
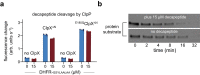
AAA+ proteases degrade intracellular proteins in a highly specific manner. E. coli ClpXP, for example, relies on a C-terminal ssrA tag or other terminal degron sequences to recognize proteins, which are then unfolded by ClpX and subsequently translocated through its axial channel and into the degradation chamber of ClpP for proteolysis. Prior cryo-EM structures reveal that the ssrA tag initially binds to a ClpX conformation in which the axial channel is closed by a pore-2 loop. Here, we show that substrate-free ClpXP has a nearly identical closed-channel conformation. We destabilize this closed-channel conformation by deleting residues from the ClpX pore-2 loop. Strikingly, open-channel ClpXP variants degrade non-native proteins lacking degrons faster than the parental enzymes in vitro but degraded GFP-ssrA more slowly. When expressed in E. coli, these open channel variants behave similarly to the wild-type enzyme in assays of filamentation and phage-Mu plating but resulted in reduced growth phenotypes at elevated temperatures or when cells were exposed to sub-lethal antibiotic concentrations. Thus, channel closure is an important determinant of ClpXP degradation specificity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
The SspB adaptor drives structural changes in the AAA+ ClpXP protease during ssrA-tagged substrate delivery.Proc Natl Acad Sci U S A. 2023 Feb 7;120(6):e2219044120. doi: 10.1073/pnas.2219044120. Epub 2023 Feb 2. Proc Natl Acad Sci U S A. 2023. PMID: 36730206 Free PMC article.
-
Multistep substrate binding and engagement by the AAA+ ClpXP protease.Proc Natl Acad Sci U S A. 2020 Nov 10;117(45):28005-28013. doi: 10.1073/pnas.2010804117. Epub 2020 Oct 26. Proc Natl Acad Sci U S A. 2020. PMID: 33106413 Free PMC article.
-
Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.Elife. 2020 Feb 28;9:e52774. doi: 10.7554/eLife.52774. Elife. 2020. PMID: 32108573 Free PMC article.
-
ClpP: a structurally dynamic protease regulated by AAA+ proteins.J Struct Biol. 2012 Aug;179(2):202-10. doi: 10.1016/j.jsb.2012.05.003. Epub 2012 May 14. J Struct Biol. 2012. PMID: 22595189 Review.
-
Structure and function of ClpXP, a AAA+ proteolytic machine powered by probabilistic ATP hydrolysis.Crit Rev Biochem Mol Biol. 2022 Apr;57(2):188-204. doi: 10.1080/10409238.2021.1979461. Epub 2021 Dec 19. Crit Rev Biochem Mol Biol. 2022. PMID: 34923891 Free PMC article. Review.
Cited by
-
A proteolytic AAA+ machine poised to unfold a protein substrate.bioRxiv [Preprint]. 2023 Dec 15:2023.12.14.571662. doi: 10.1101/2023.12.14.571662. bioRxiv. 2023. Update in: Nat Commun. 2024 Nov 8;15(1):9681. doi: 10.1038/s41467-024-53681-9 PMID: 38168193 Free PMC article. Updated. Preprint.
-
Knockout Mouse Studies Show That Mitochondrial CLPP Peptidase and CLPX Unfoldase Act in Matrix Condensates near IMM, as Fast Stress Response in Protein Assemblies for Transcript Processing, Translation, and Heme Production.Genes (Basel). 2024 May 27;15(6):694. doi: 10.3390/genes15060694. Genes (Basel). 2024. PMID: 38927630 Free PMC article. Review.
-
Automated model-free analysis of cryo-EM volume ensembles with SIREn.bioRxiv [Preprint]. 2024 Oct 8:2024.10.08.617123. doi: 10.1101/2024.10.08.617123. bioRxiv. 2024. PMID: 39415986 Free PMC article. Preprint.
-
A proteolytic AAA+ machine poised to unfold protein substrates.Nat Commun. 2024 Nov 8;15(1):9681. doi: 10.1038/s41467-024-53681-9. Nat Commun. 2024. PMID: 39516482 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

