APP-C31: An Intracellular Promoter of Both Metal-Free and Metal-Bound Amyloid-β40 Aggregation and Toxicity in Alzheimer's Disease
- PMID: 37949680
- PMCID: PMC10811509
- DOI: 10.1002/advs.202307182
APP-C31: An Intracellular Promoter of Both Metal-Free and Metal-Bound Amyloid-β40 Aggregation and Toxicity in Alzheimer's Disease
Abstract
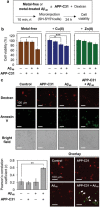
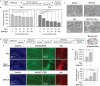
Intracellular C-terminal cleavage of the amyloid precursor protein (APP) is elevated in the brains of Alzheimer's disease (AD) patients and produces a peptide labeled APP-C31 that is suspected to be involved in the pathology of AD. But details about the role of APP-C31 in the development of the disease are not known. Here, this work reports that APP-C31 directly interacts with the N-terminal and self-recognition regions of amyloid-β40 (Aβ40 ) to form transient adducts, which facilitates the aggregation of both metal-free and metal-bound Aβ40 peptides and aggravates their toxicity. Specifically, APP-C31 increases the perinuclear and intranuclear generation of large Aβ40 deposits and, consequently, damages the nucleus leading to apoptosis. The Aβ40 -induced degeneration of neurites and inflammation are also intensified by APP-C31 in human neurons and murine brains. This study demonstrates a new function of APP-C31 as an intracellular promoter of Aβ40 amyloidogenesis in both metal-free and metal-present environments, and may offer an interesting alternative target for developing treatments for AD that have not been considered thus far.
Keywords: accelerator toward amyloidogenesis; amyloid precursor protein; amyloid-β; metal ions; protein-protein interaction.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Caspase cleavage of the amyloid precursor protein modulates amyloid beta-protein toxicity.J Neurochem. 2003 Nov;87(3):733-41. doi: 10.1046/j.1471-4159.2003.02059.x. J Neurochem. 2003. PMID: 14535955
-
The caspase-derived C-terminal fragment of betaAPP induces caspase-independent toxicity and triggers selective increase of Abeta42 in mammalian cells.J Neurochem. 2001 Sep;78(5):1153-61. doi: 10.1046/j.1471-4159.2001.00513.x. J Neurochem. 2001. PMID: 11553689
-
beta-Secretase cleavage of the amyloid precursor protein mediates neuronal apoptosis caused by familial Alzheimer's disease mutations.Brain Res Mol Brain Res. 2001 Dec 16;97(1):103-13. doi: 10.1016/s0169-328x(01)00294-7. Brain Res Mol Brain Res. 2001. PMID: 11744168
-
Regulation of Alzheimer beta-amyloid precursor trafficking and metabolism.Biochim Biophys Acta. 2000 Jul 26;1502(1):44-52. doi: 10.1016/s0925-4439(00)00031-4. Biochim Biophys Acta. 2000. PMID: 10899430 Review.
-
The amyloid precursor protein: a converging point in Alzheimer's disease.Mol Neurobiol. 2022 Jul;59(7):4501-4516. doi: 10.1007/s12035-022-02863-x. Epub 2022 May 17. Mol Neurobiol. 2022. PMID: 35579846 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- NRF-2022R1A3B1077319/National Research Foundation of Korea
- NRF-2019R1A2C1004954/National Research Foundation of Korea
- NRF-2022R1A2C1011793/National Research Foundation of Korea
- NRF-2019R1C1C1010482/National Research Foundation of Korea
- NRF-2022R1I1A2068457/National Research Foundation of Korea
- CCL22061-100/National Research Council of Science & Technology
- C320000/Korea Basic Science Institute
- C330130/Korea Basic Science Institute
- C390000/Korea Basic Science Institute
- C318410/Korea Basic Science Institute
- IBS-R010-A1/Institute for Basic Science
- NRF-2019R1A6A1A10073887/Ministry of Education
- NRF-2019H1A2A1073754/Ministry of Education
- NRF-2018H1A2A1059772/Ministry of Education
LinkOut - more resources
Full Text Sources
Medical
