Five Inhibitory Receptors Display Distinct Vesicular Distributions in Murine T Cells
- PMID: 37947636
- PMCID: PMC10649679
- DOI: 10.3390/cells12212558
Five Inhibitory Receptors Display Distinct Vesicular Distributions in Murine T Cells
Abstract
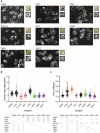
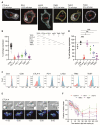
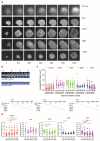
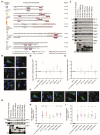
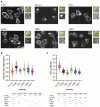
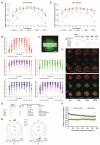
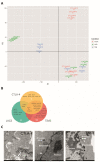
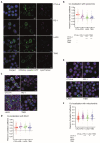
T cells can express multiple inhibitory receptors. Upon induction of T cell exhaustion in response to a persistent antigen, prominently in the anti-tumor immune response, many are expressed simultaneously. Key inhibitory receptors are CTLA-4, PD-1, LAG3, TIM3, and TIGIT, as investigated here. These receptors are important as central therapeutic targets in cancer immunotherapy. Inhibitory receptors are not constitutively expressed on the cell surface, but substantial fractions reside in intracellular vesicular structures. It remains unresolved to which extent the subcellular localization of different inhibitory receptors is distinct. Using quantitative imaging of subcellular distributions and plasma membrane insertion as complemented by proximity proteomics and biochemical analysis of the association of the inhibitory receptors with trafficking adaptors, the subcellular distributions of the five inhibitory receptors were discrete. The distribution of CTLA-4 was most distinct, with preferential association with lysosomal-derived vesicles and the sorting nexin 1/2/5/6 transport machinery. With a lack of evidence for the existence of specific vesicle subtypes to explain divergent inhibitory receptor distributions, we suggest that such distributions are driven by divergent trafficking through an overlapping joint set of vesicular structures. This extensive characterization of the subcellular localization of five inhibitory receptors in relation to each other lays the foundation for the molecular investigation of their trafficking and its therapeutic exploitation.
Keywords: T cell activation; imaging; inhibitory receptor; proximity proteomics; vesicular trafficking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Update of
-
Five inhibitory receptors display distinct vesicular distributions in T cells.bioRxiv [Preprint]. 2023 Jul 25:2023.07.21.550019. doi: 10.1101/2023.07.21.550019. bioRxiv. 2023. Update in: Cells. 2023 Oct 31;12(21):2558. doi: 10.3390/cells12212558. PMID: 37503045 Free PMC article. Updated. Preprint.
Similar articles
-
Antibodies Against Immune Checkpoint Molecules Restore Functions of Tumor-Infiltrating T Cells in Hepatocellular Carcinomas.Gastroenterology. 2017 Oct;153(4):1107-1119.e10. doi: 10.1053/j.gastro.2017.06.017. Epub 2017 Jun 23. Gastroenterology. 2017. PMID: 28648905
-
Not All Immune Checkpoints Are Created Equal.Front Immunol. 2018 Aug 31;9:1909. doi: 10.3389/fimmu.2018.01909. eCollection 2018. Front Immunol. 2018. PMID: 30233564 Free PMC article. Review.
-
TIGIT and CD96: new checkpoint receptor targets for cancer immunotherapy.Immunol Rev. 2017 Mar;276(1):112-120. doi: 10.1111/imr.12518. Immunol Rev. 2017. PMID: 28258695 Review.
-
Proteomics identification of sorting nexin 27 as a diacylglycerol kinase zeta-associated protein: new diacylglycerol kinase roles in endocytic recycling.Mol Cell Proteomics. 2007 Jun;6(6):1073-87. doi: 10.1074/mcp.M700047-MCP200. Epub 2007 Mar 9. Mol Cell Proteomics. 2007. PMID: 17351151
-
Effector Th1 cells under PD-1 and CTLA-4 checkpoint blockade abrogate the upregulation of multiple inhibitory receptors and by-pass exhaustion.Immunology. 2022 Dec;167(4):640-650. doi: 10.1111/imm.13560. Epub 2022 Aug 25. Immunology. 2022. PMID: 36053975
Cited by
-
Advancing immunotherapy for melanoma: the critical role of single-cell analysis in identifying predictive biomarkers.Front Immunol. 2024 Jul 4;15:1435187. doi: 10.3389/fimmu.2024.1435187. eCollection 2024. Front Immunol. 2024. PMID: 39026661 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

