CircBRD7 attenuates tumor growth and metastasis in nasopharyngeal carcinoma via epigenetic activation of its host gene
- PMID: 37940358
- PMCID: PMC10823269
- DOI: 10.1111/cas.15998
CircBRD7 attenuates tumor growth and metastasis in nasopharyngeal carcinoma via epigenetic activation of its host gene
Abstract
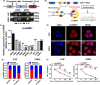
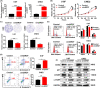
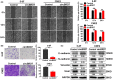
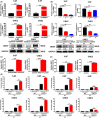
BRD7 was identified as a tumor suppressor in nasopharyngeal carcinoma (NPC). Circular RNAs (CircRNAs) are involved in the occurrence and development of NPC as oncogenes or tumor suppressors. However, the function and mechanism of the circular RNA forms derived from BRD7 in NPC are not well understood. In this study, we first identified that circBRD7 was a novel circRNA derived from BRD7 that inhibited cell proliferation, migration, invasion of NPC cells, as well as the xenograft tumor growth and metastasis in vivo. Mechanistically, circBRD7 promoted the transcriptional activation and expression of BRD7 by enhancing the enrichment of histone 3 lysine 27 acetylation (H3K27ac) in the promoter region of its host gene BRD7, and BRD7 promoted the formation of circBRD7. Therefore, circBRD7 formed a positive feedback loop with BRD7 to inhibit NPC development and progression. Moreover, restoration of BRD7 expression rescued the inhibitory effect of circBRD7 on the malignant progression of NPC. In addition, circBRD7 demonstrated low expression in NPC tissues, which was positively correlated with BRD7 expression and negatively correlated with the clinical stage of NPC patients. Taken together, circBRD7 attenuates the tumor growth and metastasis of NPC by forming a positive feedback loop with its host gene BRD7, and targeting the circBRD7/BRD7 axis is a promising strategy for the clinical diagnosis and treatment of NPC.
Keywords: BRD7; circBRD7; host gene; nasopharyngeal carcinoma; tumor progression.
© 2023 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
The authors have no conflict of interest.
Figures
Similar articles
-
BRD7 inhibits enhancer activity and expression of BIRC2 to suppress tumor growth and metastasis in nasopharyngeal carcinoma.Cell Death Dis. 2023 Feb 14;14(2):121. doi: 10.1038/s41419-023-05632-3. Cell Death Dis. 2023. PMID: 36788209 Free PMC article.
-
miR-141 is involved in BRD7-mediated cell proliferation and tumor formation through suppression of the PTEN/AKT pathway in nasopharyngeal carcinoma.Cell Death Dis. 2016 Mar 24;7(3):e2156. doi: 10.1038/cddis.2016.64. Cell Death Dis. 2016. PMID: 27010857 Free PMC article.
-
BRD7 expression and c-Myc activation forms a double-negative feedback loop that controls the cell proliferation and tumor growth of nasopharyngeal carcinoma by targeting oncogenic miR-141.J Exp Clin Cancer Res. 2018 Mar 20;37(1):64. doi: 10.1186/s13046-018-0734-2. J Exp Clin Cancer Res. 2018. PMID: 29559001 Free PMC article.
-
Promoter methylation inhibits BRD7 expression in human nasopharyngeal carcinoma cells.BMC Cancer. 2008 Sep 8;8:253. doi: 10.1186/1471-2407-8-253. BMC Cancer. 2008. PMID: 18778484 Free PMC article.
-
BRD7, a novel bromodomain gene, inhibits G1-S progression by transcriptionally regulating some important molecules involved in ras/MEK/ERK and Rb/E2F pathways.J Cell Physiol. 2004 Jul;200(1):89-98. doi: 10.1002/jcp.20013. J Cell Physiol. 2004. PMID: 15137061
Cited by
-
Back to the Origin: Mechanisms of circRNA-Directed Regulation of Host Genes in Human Disease.Noncoding RNA. 2024 Sep 24;10(5):49. doi: 10.3390/ncrna10050049. Noncoding RNA. 2024. PMID: 39452835 Free PMC article. Review.
-
hsa_circ_0072309 Inhibits Oncogenesis in Hepatocellular Carcinoma by Epigenetic Activation of its Host Gene.Cell Biochem Biophys. 2024 Dec;82(4):3251-3263. doi: 10.1007/s12013-024-01330-9. Epub 2024 Sep 16. Cell Biochem Biophys. 2024. PMID: 39283585
-
Emerging roles of circular RNAs in nasopharyngeal carcinoma: functions and implications.Cell Death Discov. 2024 Apr 25;10(1):192. doi: 10.1038/s41420-024-01964-x. Cell Death Discov. 2024. PMID: 38664370 Free PMC article. Review.
-
Hsa_circ_0092856 Promoted the Proliferation, Migration, and Invasion of NSCLC Cells by Up-Regulating the Expression of eIF3a.Biomedicines. 2024 Jan 22;12(1):247. doi: 10.3390/biomedicines12010247. Biomedicines. 2024. PMID: 38275418 Free PMC article.
References
-
- Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64‐80. - PubMed
-
- Campion NJ, Ally M, Jank BJ, Ahmed J, Alusi G. The molecular march of primary and recurrent nasopharyngeal carcinoma. Oncogene. 2021;40(10):1757‐1774. - PubMed
-
- Yu Y, Xie Y, Cao L, et al. Isolation of tumor differentially expressed genes by mixing probes library screen. Chin J Cancer. 2001;2:4‐82.
-
- Yu Y, Zhang BC, Xie Y, et al. Analysis and molecular cloning of differentially expressing genes in nasopharyngeal carcinoma. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai). 2000;32(4):327‐332. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

